DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon
- PMID: 17936300
- PMCID: PMC2121658
- DOI: 10.1016/j.jmb.2007.09.047
DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon
Abstract
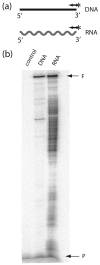
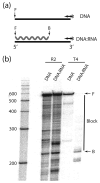
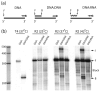
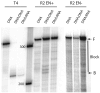
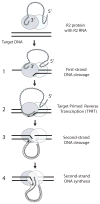
R2 elements are non-long terminal repeat (non-LTR) retrotransposons with a single open reading-frame encoding reverse transcriptase, DNA endonuclease and nucleic acid-binding domains. The elements are specialized for insertion into the 28 S rRNA genes of many animal phyla. The R2-encoded activities initiate retrotransposition by sequence-specific cleavage of the 28 S gene target site and the utilization of the released DNA 3' end to prime reverse transcription (target primed reverse transcription). The activity of the R2 polymerase on RNA templates has been shown to differ from retroviral reverse transcriptases (RTs) in a number of properties. We demonstrate that the R2-RT is capable of efficiently utilizing single-stranded DNA (ssDNA) as a template. The processivity of the enzyme on ssDNA templates is higher than its processivity on RNA templates. This finding suggests that R2-RT is also capable of synthesizing the second DNA strand during retrotransposition. However, R2-RT lacks the RNAse H activity that is typically used by retroviral and LTR-retrotransposon RTs to remove the RNA strand before the first DNA strand is used as template. Remarkably, R2-RT can displace RNA strands that are annealed to ssDNA templates with essentially no loss of processivity. Such strand displacement activity is highly unusual for a DNA polymerase. Thus the single R2 protein contains all the activities needed to make a double-stranded DNA product from an RNA transcript. Finally, during these studies we found an unexpected property of the highly sequence-specific R2 endonuclease domain. The endonuclease can non-specifically cleave ssDNA at a junction with double-stranded DNA. This activity suggests that second-strand cleavage of the target site may not be sequence specific, but rather is specified by a single-stranded region generated when the first DNA strand is used to prime reverse transcription.
Figures



Similar articles
-
The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates.J Mol Biol. 2002 Feb 22;316(3):459-73. doi: 10.1006/jmbi.2001.5369. J Mol Biol. 2002. PMID: 11866511
-
Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase.Mol Cell Biol. 1996 Sep;16(9):4726-34. doi: 10.1128/MCB.16.9.4726. Mol Cell Biol. 1996. PMID: 8756630 Free PMC article.
-
RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element.Mol Cell Biol. 1995 Jul;15(7):3882-91. doi: 10.1128/MCB.15.7.3882. Mol Cell Biol. 1995. PMID: 7540721 Free PMC article.
-
Reverse transcription of retroviruses and LTR retrotransposons.Cell Mol Life Sci. 2001 Aug;58(9):1246-62. doi: 10.1007/PL00000937. Cell Mol Life Sci. 2001. PMID: 11577982 Free PMC article. Review.
-
The diversity of retrotransposons and the properties of their reverse transcriptases.Virus Res. 2008 Jun;134(1-2):221-34. doi: 10.1016/j.virusres.2007.12.010. Epub 2008 Feb 7. Virus Res. 2008. PMID: 18261821 Free PMC article. Review.
Cited by
-
Endonuclease domain of the Drosophila melanogaster R2 non-LTR retrotransposon and related retroelements: a new model for transposition.Front Genet. 2013 Apr 26;4:63. doi: 10.3389/fgene.2013.00063. eCollection 2013. Front Genet. 2013. PMID: 23637706 Free PMC article.
-
Deamination-independent restriction of LINE-1 retrotransposition by APOBEC3H.Sci Rep. 2017 Sep 7;7(1):10881. doi: 10.1038/s41598-017-11344-4. Sci Rep. 2017. PMID: 28883657 Free PMC article.
-
Novel ribozymes: discovery, catalytic mechanisms, and the quest to understand biological function.Nucleic Acids Res. 2019 Oct 10;47(18):9480-9494. doi: 10.1093/nar/gkz737. Nucleic Acids Res. 2019. PMID: 31504786 Free PMC article.
-
Transposable elements in Drosophila.Mob Genet Elements. 2017 Apr 19;7(3):1-18. doi: 10.1080/2159256X.2017.1318201. eCollection 2017. Mob Genet Elements. 2017. PMID: 28580197 Free PMC article. Review.
-
LINE-1 ORF1 protein enhances Alu SINE retrotransposition.Gene. 2008 Aug 1;419(1-2):1-6. doi: 10.1016/j.gene.2008.04.007. Epub 2008 Apr 24. Gene. 2008. PMID: 18534786 Free PMC article.
References
-
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. - PubMed
-
- Whitcomb JM, Hughes SH. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. - PubMed
-
- Voytas DF, Boeke JD. Ty1 and Ty5 of Sacharomyces cerevisiae. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. American Society for Microbiology; Washington DC: 2002. pp. 631–662.
-
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. - PubMed
-
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

