Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression
- PMID: 17936713
- PMCID: PMC2034406
- DOI: 10.1016/j.molcel.2007.07.030
Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression
Abstract
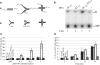
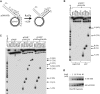
Lesions in the template DNA strand block the progression of the replication fork. In the yeast Saccharomyces cerevisiae, replication through DNA lesions is mediated by different Rad6-Rad18-dependent means, which include translesion synthesis and a Rad5-dependent postreplicational repair pathway that repairs the discontinuities that form in the DNA synthesized from damaged templates. Although translesion synthesis is well characterized, little is known about the mechanisms that modulate Rad5-dependent postreplicational repair. Here we show that yeast Rad5 has a DNA helicase activity that is specialized for replication fork regression. On model replication fork structures, Rad5 concertedly unwinds and anneals the nascent and the parental strands without exposing extended single-stranded regions. These observations provide insight into the mechanism of postreplicational repair in which Rad5 action promotes template switching for error-free damage bypass.
Figures

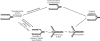
Comment in
-
Reversal of fortune: Rad5 to the rescue.Mol Cell. 2007 Oct 26;28(2):181-3. doi: 10.1016/j.molcel.2007.10.001. Mol Cell. 2007. PMID: 17964257
Similar articles
-
Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Apr;22(7):2419-26. doi: 10.1128/MCB.22.7.2419-2426.2002. Mol Cell Biol. 2002. PMID: 11884624 Free PMC article.
-
Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae.Mol Cell Biol. 2006 Oct;26(20):7783-90. doi: 10.1128/MCB.01260-06. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908531 Free PMC article.
-
Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Nov;27(21):7758-64. doi: 10.1128/MCB.01331-07. Epub 2007 Sep 4. Mol Cell Biol. 2007. PMID: 17785441 Free PMC article.
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
-
The DNA damage tolerance factor Rad5 and telomere replication.Curr Genet. 2025 May 26;71(1):11. doi: 10.1007/s00294-025-01315-y. Curr Genet. 2025. PMID: 40418329 Free PMC article. Review.
Cited by
-
Characterization of human Spartan/C1orf124, an ubiquitin-PCNA interacting regulator of DNA damage tolerance.Nucleic Acids Res. 2012 Nov;40(21):10795-808. doi: 10.1093/nar/gks850. Epub 2012 Sep 16. Nucleic Acids Res. 2012. PMID: 22987070 Free PMC article.
-
Cyclin-dependent kinase modulates budding yeast Rad5 stability during cell cycle.PLoS One. 2018 Sep 26;13(9):e0204680. doi: 10.1371/journal.pone.0204680. eCollection 2018. PLoS One. 2018. PMID: 30256854 Free PMC article.
-
Saccharomyces cerevisiae Mus81-Mms4 prevents accelerated senescence in telomerase-deficient cells.PLoS Genet. 2020 May 29;16(5):e1008816. doi: 10.1371/journal.pgen.1008816. eCollection 2020 May. PLoS Genet. 2020. PMID: 32469862 Free PMC article.
-
Post-Translational Modifications of PCNA: Guiding for the Best DNA Damage Tolerance Choice.J Fungi (Basel). 2022 Jun 10;8(6):621. doi: 10.3390/jof8060621. J Fungi (Basel). 2022. PMID: 35736104 Free PMC article. Review.
-
Histone H3 K79 methylation states play distinct roles in UV-induced sister chromatid exchange and cell cycle checkpoint arrest in Saccharomyces cerevisiae.Nucleic Acids Res. 2014 Jun;42(10):6286-99. doi: 10.1093/nar/gku242. Epub 2014 Apr 19. Nucleic Acids Res. 2014. PMID: 24748660 Free PMC article.
References
-
- Bailly V., Lamb J., Sung P., Prakash S., Prakash L. Specific complex formation between yeast Rad6 and Rad18 proteins—a potential mechanism for targeting Rad6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 1994;8:811–820. - PubMed
-
- Bailly V., Lauder S., Prakash S., Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 1997;272:23360–23365. - PubMed
-
- Cordeiro-Stone M., Zaritskaya L.S., Price L.K., Kaufmann W.K. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem. 1997;272:13945–13954. - PubMed
-
- Cordeiro-Stone M., Makhov A.M., Zaritskaya L.S., Griffith J.D. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 1999;289:1207–1218. - PubMed
-
- Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopes M., Sogo J., Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

