CREB binding and activity in brain: regional specificity and induction by electroconvulsive seizure
- PMID: 17936724
- PMCID: PMC3691692
- DOI: 10.1016/j.biopsych.2007.08.003
CREB binding and activity in brain: regional specificity and induction by electroconvulsive seizure
Abstract
Background: The transcription factor cyclic adenosine monophosphate response element binding protein (CREB) orchestrates diverse neurobiological processes including cell differentiation, survival, and plasticity. Alterations in CREB-mediated transcription have been implicated in numerous central nervous system (CNS) disorders including depression, anxiety, addiction, and cognitive decline. However, it remains unclear how CREB contributes to normal and aberrant CNS function, as the identity of CREB-regulated genes in brain and the regional and temporal dynamics of CREB function remain largely undetermined.
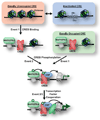
Methods: We combined microarray and chromatin immunoprecipitation technology to analyze CREB-DNA interactions in brain. We compared the occupancy and activity of CREB at gene promoters in rat frontal cortex, hippocampus, and striatum before and after a rodent model of electroconvulsive therapy.
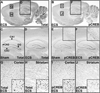
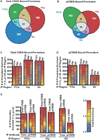
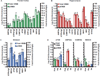
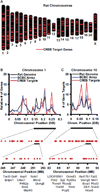
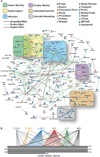
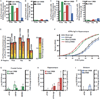
Results: Our analysis identified >860 CREB binding sites in rat brain. We identified multiple genomic loci enriched with CREB binding sites and find that CREB-occupied transcripts interact extensively to promote cell proliferation, plasticity, and resiliency. We discovered regional differences in CREB occupancy and activity that explain, in part, the diverse biological and behavioral outputs of CREB activity in frontal cortex, hippocampus, and striatum. Electroconvulsive seizure rapidly increased CREB occupancy and/or phosphorylation at select promoters, demonstrating that both events contribute to the temporal regulation of the CREB transcriptome.
Conclusions: Our data provide a mechanistic basis for CREB's ability to integrate regional and temporal cues to orchestrate state-specific patterns of transcription in the brain, indicate that CREB is an important mediator of the biological responses to electroconvulsive seizure, and provide global mechanistic insights into CREB's role in psychiatric and cognitive function.
Conflict of interest statement
Dr. Tanis reported no conflicts of interest pertaining to this work. Dr. Duman reported no conflicts of interest pertaining to this work. Dr. Newton reported no conflicts of interest pertaining to this work.
Figures


Similar articles
-
Analysis of target genes regulated by chronic electroconvulsive therapy reveals role for Fzd6 in depression.Biol Psychiatry. 2012 Jan 1;71(1):51-8. doi: 10.1016/j.biopsych.2011.08.004. Epub 2011 Sep 19. Biol Psychiatry. 2012. PMID: 21937024 Free PMC article.
-
Alterations in long-term seizure susceptibility and the complex of PSD-95 with NMDA receptor from animals previously exposed to perinatal hypoxia.Epilepsia. 2006 Feb;47(2):288-96. doi: 10.1111/j.1528-1167.2006.00420.x. Epilepsia. 2006. PMID: 16499752
-
Neuronal activity increases the phosphorylation of the transcription factor cAMP response element-binding protein (CREB) in rat hippocampus and cortex.J Biol Chem. 1996 Jun 14;271(24):14214-20. doi: 10.1074/jbc.271.24.14214. J Biol Chem. 1996. PMID: 8662977
-
CREB's control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models.Trends Neurosci. 2010 May;33(5):230-40. doi: 10.1016/j.tins.2010.02.001. Epub 2010 Mar 10. Trends Neurosci. 2010. PMID: 20223527 Review.
-
The involvement of ERK/CREB/Bcl-2 in depression-like behavior in prenatally stressed offspring rats.Brain Res Bull. 2013 Oct;99:1-8. doi: 10.1016/j.brainresbull.2013.08.003. Epub 2013 Sep 1. Brain Res Bull. 2013. PMID: 24004471 Review.
Cited by
-
Increased Expression of Brain-Derived Neurotrophic Factor Transcripts I and VI, cAMP Response Element Binding, and Glucocorticoid Receptor in the Cortex of Patients with Temporal Lobe Epilepsy.Mol Neurobiol. 2018 May;55(5):3698-3708. doi: 10.1007/s12035-017-0597-0. Epub 2017 May 19. Mol Neurobiol. 2018. PMID: 28527108
-
Delta FosB-mediated alterations in dopamine signaling are normalized by a palatable high-fat diet.Biol Psychiatry. 2008 Dec 1;64(11):941-50. doi: 10.1016/j.biopsych.2008.06.007. Epub 2008 Jul 26. Biol Psychiatry. 2008. PMID: 18657800 Free PMC article.
-
Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins.Neuron. 2009 May 14;62(3):335-48. doi: 10.1016/j.neuron.2009.03.026. Neuron. 2009. PMID: 19447090 Free PMC article.
-
Yohimbine increases impulsivity through activation of cAMP response element binding in the orbitofrontal cortex.Biol Psychiatry. 2010 Apr 1;67(7):649-56. doi: 10.1016/j.biopsych.2009.11.030. Epub 2010 Feb 16. Biol Psychiatry. 2010. PMID: 20163788 Free PMC article.
-
Hippocampal glucocorticoid target genes associated with enhancement of memory consolidation.Eur J Neurosci. 2022 May;55(9-10):2666-2683. doi: 10.1111/ejn.15226. Epub 2021 May 6. Eur J Neurosci. 2022. PMID: 33840130 Free PMC article.
References
-
- Shaywitz A, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. - PubMed
-
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. - PubMed
-
- Carlezon WA, Jr., Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. - PubMed
-
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. - PubMed

