Animal protection and structural studies of a consensus sequence vaccine targeting the receptor binding domain of the type IV pilus of Pseudomonas aeruginosa
- PMID: 17936788
- PMCID: PMC3493149
- DOI: 10.1016/j.jmb.2007.09.032
Animal protection and structural studies of a consensus sequence vaccine targeting the receptor binding domain of the type IV pilus of Pseudomonas aeruginosa
Abstract
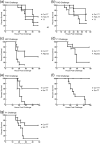
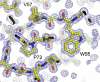
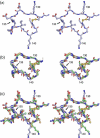
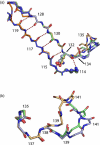
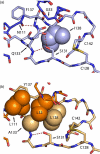
One of the main obstacles in the development of a vaccine against Pseudomonas aeruginosa is the requirement that it is protective against a wide range of virulent strains. We have developed a synthetic-peptide consensus-sequence vaccine (Cs1) that targets the host receptor-binding domain (RBD) of the type IV pilus of P. aeruginosa. Here, we show that this vaccine provides increased protection against challenge by the four piliated strains that we have examined (PAK, PAO, KB7 and P1) in the A.BY/SnJ mouse model of acute P. aeruginosa infection. To further characterize the consensus sequence, we engineered Cs1 into the PAK monomeric pilin protein and determined the crystal structure of the chimeric Cs1 pilin to 1.35 A resolution. The substitutions (T130K and E135P) used to create Cs1 do not disrupt the conserved backbone conformation of the pilin RBD. In fact, based on the Cs1 pilin structure, we hypothesize that the E135P substitution bolsters the conserved backbone conformation and may partially explain the immunological activity of Cs1. Structural analysis of Cs1, PAK and K122-4 pilins reveal substitutions of non-conserved residues in the RBD are compensated for by complementary changes in the rest of the pilin monomer. Thus, the interactions between the RBD and the rest of the pilin can either be mediated by polar interactions of a hydrogen bond network in some strains or by hydrophobic interactions in others. Both configurations maintain a conserved backbone conformation of the RBD. Thus, the backbone conformation is critical in our consensus-sequence vaccine design and that cross-reactivity of the antibody response may be modulated by the composition of exposed side-chains on the surface of the RBD. This structure will guide our future vaccine design by focusing our investigation on the four variable residue positions that are exposed on the RBD surface.
Figures
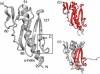

Similar articles
-
Interaction of the receptor binding domains of Pseudomonas aeruginosa pili strains PAK, PAO, KB7 and P1 to a cross-reactive antibody and receptor analog: implications for synthetic vaccine design.J Mol Biol. 1997 Mar 28;267(2):382-402. doi: 10.1006/jmbi.1996.0871. J Mol Biol. 1997. PMID: 9096233
-
Backbone dynamics of receptor binding and antigenic regions of a Pseudomonas aeruginosa pilin monomer.Biochemistry. 2001 Apr 3;40(13):3985-95. doi: 10.1021/bi002524h. Biochemistry. 2001. PMID: 11300779
-
Interaction of a bacterially expressed peptide from the receptor binding domain of Pseudomonas aeruginosa pili strain PAK with a cross-reactive antibody: conformation of the bound peptide.Biochemistry. 2000 Dec 5;39(48):14847-64. doi: 10.1021/bi0016568. Biochemistry. 2000. PMID: 11101301
-
The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa--a review.Gene. 1997 Jun 11;192(1):99-108. doi: 10.1016/s0378-1119(97)00116-9. Gene. 1997. PMID: 9224879 Review.
-
Pseudomonas aeruginosa antigens as potential vaccines.FEMS Microbiol Rev. 1997 Nov;21(3):243-77. doi: 10.1111/j.1574-6976.1997.tb00353.x. FEMS Microbiol Rev. 1997. PMID: 9451816 Review.
Cited by
-
Fibril-mediated oligomerization of pilin-derived protein nanotubes.J Nanobiotechnology. 2013 Jul 5;11:24. doi: 10.1186/1477-3155-11-24. J Nanobiotechnology. 2013. PMID: 23829476 Free PMC article.
-
Dimerization of the type IV pilin from Pseudomonas aeruginosa strain K122-4 results in increased helix stability as measured by time-resolved hydrogen-deuterium exchange.Struct Dyn. 2015 Aug 28;3(1):012001. doi: 10.1063/1.4929597. eCollection 2016 Jan. Struct Dyn. 2015. PMID: 26798830 Free PMC article.
-
Construction, expression, purification and characterization of secretin domain of PilQ and triple PilA-related disulfide loop peptides fusion protein from Pseudomonas aeruginosa.Iran J Basic Med Sci. 2017 May;20(5):458-466. doi: 10.22038/IJBMS.2017.8667. Iran J Basic Med Sci. 2017. PMID: 28656079 Free PMC article.
-
Vaccination to Prevent Pseudomonas aeruginosa Bloodstream Infections.Front Microbiol. 2022 Mar 28;13:870104. doi: 10.3389/fmicb.2022.870104. eCollection 2022. Front Microbiol. 2022. PMID: 35418967 Free PMC article.
-
Peptide-based synthetic vaccines.Chem Sci. 2016 Feb 1;7(2):842-854. doi: 10.1039/c5sc03892h. Epub 2015 Dec 17. Chem Sci. 2016. PMID: 28791117 Free PMC article. Review.
References
-
- Braunwald E. Harrison's Principles of Internal Medicine. 15th edit. McGraw-Hill; New York: 2001.
-
- Davies JC. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr. Respir. Rev. 2002;3:128–134. - PubMed
-
- Paranchych W, Sastry PA, Volpel K, Loh BA, Speert DP. Fimbriae (pili): Molecular basis of Pseudomonas aeruginosa adherence. Clin. Invest. Med. 1986;9:113–118. - PubMed
-
- Mattick J. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002;56:289–314. - PubMed
-
- Hazes B, Sastry PA, Hayakawa K, Read RJ, Irvin RT. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 2000;299:1005–1017. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

