Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection
- PMID: 17937498
- PMCID: PMC2014795
- DOI: 10.1371/journal.ppat.0030144
Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection
Abstract
Despite the importance of vaccinia virus in basic and applied immunology, our knowledge of the human immune response directed against this virus is very limited. CD4(+) T cell responses are an important component of immunity induced by current vaccinia-based vaccines, and likely will be required for new subunit vaccine approaches, but to date vaccinia-specific CD4(+) T cell responses have been poorly characterized, and CD4(+) T cell epitopes have been reported only recently. Classical approaches used to identify T cell epitopes are not practical for large genomes like vaccinia. We developed and validated a highly efficient computational approach that combines prediction of class II MHC-peptide binding activity with prediction of antigen processing and presentation. Using this approach and screening only 36 peptides, we identified 25 epitopes recognized by T cells from vaccinia-immune individuals. Although the predictions were made for HLA-DR1, eight of the peptides were recognized by donors of multiple haplotypes. T cell responses were observed in samples of peripheral blood obtained many years after primary vaccination, and were amplified after booster immunization. Peptides recognized by multiple donors are highly conserved across the poxvirus family, including variola, the causative agent of smallpox, and may be useful in development of a new generation of smallpox vaccines and in the analysis of the immune response elicited to vaccinia virus. Moreover, the epitope identification approach developed here should find application to other large-genome pathogens.
Conflict of interest statement
Figures


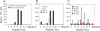
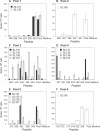
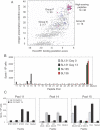
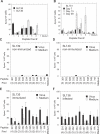
Similar articles
-
Human cytotoxic CD4+ T cells recognize HLA-DR1-restricted epitopes on vaccinia virus proteins A24R and D1R conserved among poxviruses.J Immunol. 2007 Jul 15;179(2):1303-12. doi: 10.4049/jimmunol.179.2.1303. J Immunol. 2007. PMID: 17617623
-
Vaccinia peptides eluted from HLA-DR1 isolated from virus-infected cells are recognized by CD4+ T cells from a vaccinated donor.J Proteome Res. 2008 Jul;7(7):2703-11. doi: 10.1021/pr700780x. Epub 2008 May 29. J Proteome Res. 2008. PMID: 18507432 Free PMC article.
-
Identification of protective T-cell antigens for smallpox vaccines.Cytotherapy. 2020 Nov;22(11):642-652. doi: 10.1016/j.jcyt.2020.04.098. Epub 2020 May 8. Cytotherapy. 2020. PMID: 32747299 Free PMC article.
-
T-Cell epitope discovery for variola and vaccinia viruses.Rev Med Virol. 2007 Mar-Apr;17(2):93-113. doi: 10.1002/rmv.527. Rev Med Virol. 2007. PMID: 17195963 Review.
-
Definition of epitopes and antigens recognized by vaccinia specific immune responses: their conservation in variola virus sequences, and use as a model system to study complex pathogens.Vaccine. 2009 Dec 30;27 Suppl 6(Suppl 6):G21-6. doi: 10.1016/j.vaccine.2009.10.011. Vaccine. 2009. PMID: 20006135 Free PMC article. Review.
Cited by
-
CD4+ T cells provide intermolecular help to generate robust antibody responses in vaccinia virus-vaccinated humans.J Immunol. 2013 Jun 15;190(12):6023-33. doi: 10.4049/jimmunol.1202523. Epub 2013 May 10. J Immunol. 2013. PMID: 23667112 Free PMC article.
-
A novel method to measure HLA-DM-susceptibility of peptides bound to MHC class II molecules based on peptide binding competition assay and differential IC(50) determination.J Immunol Methods. 2014 Apr;406:21-33. doi: 10.1016/j.jim.2014.02.008. Epub 2014 Feb 25. J Immunol Methods. 2014. PMID: 24583195 Free PMC article.
-
Circulating human rotavirus specific CD4 T cells identified with a class II tetramer express the intestinal homing receptors α4β7 and CCR9.Virology. 2014 Mar;452-453:191-201. doi: 10.1016/j.virol.2014.01.014. Epub 2014 Feb 7. Virology. 2014. PMID: 24606696 Free PMC article.
-
Citrullination only infrequently impacts peptide binding to HLA class II MHC.PLoS One. 2017 May 8;12(5):e0177140. doi: 10.1371/journal.pone.0177140. eCollection 2017. PLoS One. 2017. PMID: 28481943 Free PMC article.
-
GM-CSF production allows the identification of immunoprevalent antigens recognized by human CD4+ T cells following smallpox vaccination.PLoS One. 2011;6(9):e24091. doi: 10.1371/journal.pone.0024091. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931646 Free PMC article.
References
-
- Amanna IJ, Slifka MK, Crotty S. Immunity and immunological memory following smallpox vaccination. Immunol Rev. 2006;211:320–337. - PubMed
-
- Mack TM, Noble J, Jr, Thomas DB. A prospective study of serum antibody and protection against smallpox. Am J Trop Med Hyg. 1972;21:214–218. - PubMed
-
- Edghill-Smith Y, Golding H, Manischewitz J, King LR, Scott D, et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11:740–747. - PubMed
-
- Fang M, Sigal LJ. Antibodies and CD8+ T cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J Immunol. 2005;175:6829–6836. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

