Large scale statistical inference of signaling pathways from RNAi and microarray data
- PMID: 17937790
- PMCID: PMC2241646
- DOI: 10.1186/1471-2105-8-386
Large scale statistical inference of signaling pathways from RNAi and microarray data
Abstract
Background: The advent of RNA interference techniques enables the selective silencing of biologically interesting genes in an efficient way. In combination with DNA microarray technology this enables researchers to gain insights into signaling pathways by observing downstream effects of individual knock-downs on gene expression. These secondary effects can be used to computationally reverse engineer features of the upstream signaling pathway.
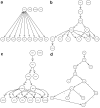
Results: In this paper we address this challenging problem by extending previous work by Markowetz et al., who proposed a statistical framework to score networks hypotheses in a Bayesian manner. Our extensions go in three directions: First, we introduce a way to omit the data discretization step needed in the original framework via a calculation based on p-values instead. Second, we show how prior assumptions on the network structure can be incorporated into the scoring scheme using regularization techniques. Third and most important, we propose methods to scale up the original approach, which is limited to around 5 genes, to large scale networks.
Conclusion: Comparisons of these methods on artificial data are conducted. Our proposed module network is employed to infer the signaling network between 13 genes in the ER-alpha pathway in human MCF-7 breast cancer cells. Using a bootstrapping approach this reconstruction can be found with good statistical stability. The code for the module network inference method is available in the latest version of the R-package nem, which can be obtained from the Bioconductor homepage.
Figures


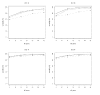
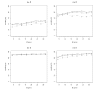
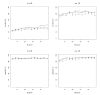
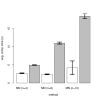

References
-
- Tikhonov A, Arsenin V. Solutions of ill-posed problems. W.H. Winston; 1977.
-
- Smyth G. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004. p. 3. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

