Distribution of DNA replication proteins in Drosophila cells
- PMID: 17937809
- PMCID: PMC2104529
- DOI: 10.1186/1471-2121-8-42
Distribution of DNA replication proteins in Drosophila cells
Abstract
Background: DNA replication in higher eukaryotic cells is organized in discrete subnuclear sites called replication foci (RF). During the S phase, most replication proteins assemble at the RF by interacting with PCNA via a PCNA binding domain (PBD). This has been shown to occur for many mammalian replication proteins, but it is not known whether this mechanism is conserved in evolution.
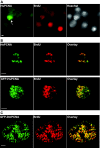
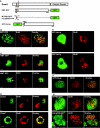
Results: Fluorescent fusions of mammalian replication proteins, Dnmt1, HsDNA Lig I and HsPCNA were analyzed for their ability to target to RF in Drosophila cells. Except for HsPCNA, none of the other proteins and their deletions showed any accumulation at RF in Drosophila cells. We hypothesized that in Drosophila cells there might be some other peptide sequence responsible for targeting proteins to RF. To test this, we identified the DmDNA Lig I and compared the protein sequence with HsDNA Lig I. The two orthologs shared the PBD suggesting a functionally conserved role for this domain in the Drosophila counterpart. A series of deletions of DmDNA Lig I were analyzed for their ability to accumulate at RF in Drosophila and mammalian cells. Surprisingly, no accumulation at RF was observed in Drosophila cells, while in mammalian cells DmDNA Lig I accumulated at RF via its PBD. Further, GFP fusions with the PBD domains from Dnmt1, HsDNA Lig I and DmDNA Lig I, were able to target to RF only in mammalian cells but not in Drosophila cells.
Conclusion: We show that S phase in Drosophila cells is characterized by formation of RF marked by PCNA like in mammalian cells. However, other than PCNA none of the replication proteins and their deletions tested here showed accumulation at RF in Drosophila cells while the same proteins and deletions are capable of accumulating at RF in mammalian cells. We hypothesize that unlike mammalian cells, in Drosophila cells, replication proteins do not form long-lasting interactions with the replication machinery, and rather perform their functions via very transient interactions at the RF.
Figures



Similar articles
-
DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories.EMBO J. 1998 Jul 1;17(13):3786-95. doi: 10.1093/emboj/17.13.3786. EMBO J. 1998. PMID: 9649448 Free PMC article.
-
A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells.EMBO J. 1996 Aug 15;15(16):4423-33. EMBO J. 1996. PMID: 8861969 Free PMC article.
-
PCNA binding proteins in Drosophila melanogaster : the analysis of a conserved PCNA binding domain.Nucleic Acids Res. 1998 Sep 1;26(17):3925-32. doi: 10.1093/nar/26.17.3925. Nucleic Acids Res. 1998. PMID: 9705499 Free PMC article.
-
DNA ligase I, the replicative DNA ligase.Subcell Biochem. 2012;62:327-41. doi: 10.1007/978-94-007-4572-8_17. Subcell Biochem. 2012. PMID: 22918593 Free PMC article. Review.
-
DNA replication: partners in the Okazaki two-step.Curr Biol. 2001 Oct 16;11(20):R842-4. doi: 10.1016/s0960-9822(01)00500-0. Curr Biol. 2001. PMID: 11676941 Review.
Cited by
-
Chromatin condensation modulates access and binding of nuclear proteins.FASEB J. 2010 Apr;24(4):1066-72. doi: 10.1096/fj.08-128959. Epub 2009 Nov 6. FASEB J. 2010. PMID: 19897663 Free PMC article.
-
PcG-mediated higher-order chromatin structures modulate replication programs at the Drosophila BX-C.PLoS Genet. 2013;9(2):e1003283. doi: 10.1371/journal.pgen.1003283. Epub 2013 Feb 21. PLoS Genet. 2013. PMID: 23437006 Free PMC article.
-
Isoform-specific and ubiquitination dependent recruitment of Tet1 to replicating heterochromatin modulates methylcytosine oxidation.Nat Commun. 2022 Sep 2;13(1):5173. doi: 10.1038/s41467-022-32799-8. Nat Commun. 2022. PMID: 36056023 Free PMC article.
-
Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus.PLoS Pathog. 2009 Mar;5(3):e1000343. doi: 10.1371/journal.ppat.1000343. Epub 2009 Mar 20. PLoS Pathog. 2009. PMID: 19300496 Free PMC article.
References
-
- Sparvoli E, Levi M, Rossi E. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci. 1994;107:3097–3103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

