Topology independent protein structural alignment
- PMID: 17937816
- PMCID: PMC2096629
- DOI: 10.1186/1471-2105-8-388
Topology independent protein structural alignment
Abstract
Background: Identifying structurally similar proteins with different chain topologies can aid studies in homology modeling, protein folding, protein design, and protein evolution. These include circular permuted protein structures, and the more general cases of non-cyclic permutations between similar structures, which are related by non-topological rearrangement beyond circular permutation. We present a method based on an approximation algorithm that finds sequence-order independent structural alignments that are close to optimal. We formulate the structural alignment problem as a special case of the maximum-weight independent set problem, and solve this computationally intensive problem approximately by iteratively solving relaxations of a corresponding integer programming problem. The resulting structural alignment is sequence order independent. Our method is also insensitive to insertions, deletions, and gaps.
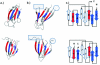
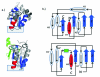
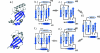
Results: Using a novel similarity score and a statistical model for significance p-value, we are able to discover previously unknown circular permuted proteins between nucleoplasmin-core protein and auxin binding protein, between aspartate rasemase and 3-dehydrogenate dehydralase, as well as between migration inhibition factor and arginine repressor which involves an additional strand-swapping. We also report the finding of non-cyclic permuted protein structures existing in nature between AML1/core binding factor and ribofalvin synthase. Our method can be used for large scale alignment of protein structures regardless of the topology.
Conclusion: The approximation algorithm introduced in this work can find good solutions for the problem of protein structure alignment. Furthermore, this algorithm can detect topological differences between two spatially similar protein structures. The alignment between MIF and the arginine repressor demonstrates our algorithm's ability to detect structural similarities even when spatial rearrangement of structural units has occurred. The effectiveness of our method is also demonstrated by the discovery of previously unknown circular permutations. In addition, we report in this study the finding of a naturally occurring non-cyclic permuted protein between AML1/Core Binding Factor chain F and riboflavin synthase chain A.
Figures

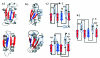
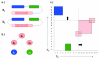
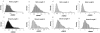
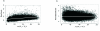

References
-
- Binkowski TA, DasGupta B, Liang J. Order independent structural alignment of circularly permuted proteins. Conf Proc IEEE Eng Med Biol Soc. 2004;4:2781–2784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

