MindSeer: a portable and extensible tool for visualization of structural and functional neuroimaging data
- PMID: 17937818
- PMCID: PMC2099449
- DOI: 10.1186/1471-2105-8-389
MindSeer: a portable and extensible tool for visualization of structural and functional neuroimaging data
Abstract
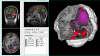
Background: Three-dimensional (3-D) visualization of multimodality neuroimaging data provides a powerful technique for viewing the relationship between structure and function. A number of applications are available that include some aspect of 3-D visualization, including both free and commercial products. These applications range from highly specific programs for a single modality, to general purpose toolkits that include many image processing functions in addition to visualization. However, few if any of these combine both stand-alone and remote multi-modality visualization in an open source, portable and extensible tool that is easy to install and use, yet can be included as a component of a larger information system.
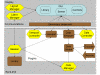
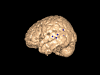
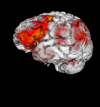
Results: We have developed a new open source multimodality 3-D visualization application, called MindSeer, that has these features: integrated and interactive 3-D volume and surface visualization, Java and Java3D for true cross-platform portability, one-click installation and startup, integrated data management to help organize large studies, extensibility through plugins, transparent remote visualization, and the ability to be integrated into larger information management systems. We describe the design and implementation of the system, as well as several case studies that demonstrate its utility. These case studies are available as tutorials or demos on the associated website: http://sig.biostr.washington.edu/projects/MindSeer.
Conclusion: MindSeer provides a powerful visualization tool for multimodality neuroimaging data. Its architecture and unique features also allow it to be extended into other visualization domains within biomedicine.
Figures



Similar articles
-
NeuroTerrain--a client-server system for browsing 3D biomedical image data sets.BMC Bioinformatics. 2007 Feb 5;8:40. doi: 10.1186/1471-2105-8-40. BMC Bioinformatics. 2007. PMID: 17280615 Free PMC article.
-
Synchronized 2D/3D optical mapping for interactive exploration and real-time visualization of multi-function neurological images.Comput Med Imaging Graph. 2013 Oct-Dec;37(7-8):552-67. doi: 10.1016/j.compmedimag.2013.06.007. Epub 2013 Aug 20. Comput Med Imaging Graph. 2013. PMID: 23968722
-
PET/CT image navigation and communication.J Nucl Med. 2004 Jan;45 Suppl 1:46S-55S. J Nucl Med. 2004. PMID: 14736835 Review.
-
Internet image viewer (iiV).BMC Med Imaging. 2008 May 29;8:10. doi: 10.1186/1471-2342-8-10. BMC Med Imaging. 2008. PMID: 18510765 Free PMC article.
-
Volume visualization: a technical overview with a focus on medical applications.J Digit Imaging. 2011 Aug;24(4):640-64. doi: 10.1007/s10278-010-9321-6. J Digit Imaging. 2011. PMID: 20714917 Free PMC article. Review.
Cited by
-
A query integrator and manager for the query web.J Biomed Inform. 2012 Oct;45(5):975-91. doi: 10.1016/j.jbi.2012.03.008. Epub 2012 Apr 17. J Biomed Inform. 2012. PMID: 22531831 Free PMC article.
-
GRACE: A visual comparison framework for integrated spatial and non-spatial geriatric data.IEEE Trans Vis Comput Graph. 2013 Dec;19(12):2916-25. doi: 10.1109/TVCG.2013.161. IEEE Trans Vis Comput Graph. 2013. PMID: 24051859 Free PMC article.
-
Neuroanatomical domain of the foundational model of anatomy ontology.J Biomed Semantics. 2014 Jan 8;5(1):1. doi: 10.1186/2041-1480-5-1. J Biomed Semantics. 2014. PMID: 24398054 Free PMC article.
-
A symmetrical Waxholm canonical mouse brain for NeuroMaps.J Neurosci Methods. 2011 Feb 15;195(2):170-5. doi: 10.1016/j.jneumeth.2010.11.028. Epub 2010 Dec 14. J Neurosci Methods. 2011. PMID: 21163300 Free PMC article.
-
Faceted visualization of three dimensional neuroanatomy by combining ontology with faceted search.Neuroinformatics. 2014 Apr;12(2):245-59. doi: 10.1007/s12021-013-9202-5. Neuroinformatics. 2014. PMID: 24006207 Free PMC article.
References
-
- Kennedy DN. Internet Analysis Tools Registry http://www.cma.mgh.harvard.edu/iatr/index.php - PubMed
-
- Pascual-Marqui RD, Esslen M, Kochi K, Lehmann D. Functional imaging with low resolution brain electromagnetic tomography (LORETA): a review. Methods and Findings in Experimental and Clinical Pharmacology. 2002;24C:91–95. - PubMed
-
- FMRIDB Image Analysis Group FSL - The FMRIB Software Libarary http://www.fmrib.ox.ac.uk/fsl/index.html
-
- Wellcome Department of Cognitive Neurology Statistical Parametric Mapping http://www.fil.ion.ucl.ac.uk/spm/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

