On the detection of functionally coherent groups of protein domains with an extension to protein annotation
- PMID: 17937820
- PMCID: PMC2151957
- DOI: 10.1186/1471-2105-8-390
On the detection of functionally coherent groups of protein domains with an extension to protein annotation
Abstract
Background: Protein domains coordinate to perform multifaceted cellular functions, and domain combinations serve as the functional building blocks of the cell. The available methods to identify functional domain combinations are limited in their scope, e.g. to the identification of combinations falling within individual proteins or within specific regions in a translated genome. Further effort is needed to identify groups of domains that span across two or more proteins and are linked by a cooperative function. Such functional domain combinations can be useful for protein annotation.
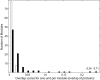
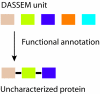
Results: Using a new computational method, we have identified 114 groups of domains, referred to as domain assembly units (DASSEM units), in the proteome of budding yeast Saccharomyces cerevisiae. The units participate in many important cellular processes such as transcription regulation, translation initiation, and mRNA splicing. Within the units the domains were found to function in a cooperative manner; and each domain contributed to a different aspect of the unit's overall function. The member domains of DASSEM units were found to be significantly enriched among proteins contained in transcription modules, defined as genes sharing similar expression profiles and presumably similar functions. The observation further confirmed the functional coherence of DASSEM units. The functional linkages of units were found in both functionally characterized and uncharacterized proteins, which enabled the assessment of protein function based on domain composition.
Conclusion: A new computational method was developed to identify groups of domains that are linked by a common function in the proteome of Saccharomyces cerevisiae. These groups can either lie within individual proteins or span across different proteins. We propose that the functional linkages among the domains within the DASSEM units can be used as a non-homology based tool to annotate uncharacterized proteins.
Figures


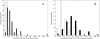

Similar articles
-
AVID: an integrative framework for discovering functional relationships among proteins.BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article.
-
Accurate extraction of functional associations between proteins based on common interaction partners and common domains.Bioinformatics. 2005 May 1;21(9):2043-8. doi: 10.1093/bioinformatics/bti305. Epub 2005 Feb 4. Bioinformatics. 2005. PMID: 15699027
-
Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins.Genome Biol. 2006;7(4):R30. doi: 10.1186/gb-2006-7-4-r30. Epub 2006 Apr 10. Genome Biol. 2006. PMID: 16606443 Free PMC article.
-
Predicting protein function from sequence and structural data.Curr Opin Struct Biol. 2005 Jun;15(3):275-84. doi: 10.1016/j.sbi.2005.04.003. Curr Opin Struct Biol. 2005. PMID: 15963890 Review.
-
The application of modular protein domains in proteomics.FEBS Lett. 2012 Aug 14;586(17):2586-96. doi: 10.1016/j.febslet.2012.04.019. Epub 2012 Apr 21. FEBS Lett. 2012. PMID: 22710164 Free PMC article. Review.
Cited by
-
CeGAL: Redefining a Widespread Fungal-Specific Transcription Factor Family Using an In Silico Error-Tracking Approach.J Fungi (Basel). 2023 Mar 29;9(4):424. doi: 10.3390/jof9040424. J Fungi (Basel). 2023. PMID: 37108879 Free PMC article.
-
Improvement in Protein Domain Identification Is Reached by Breaking Consensus, with the Agreement of Many Profiles and Domain Co-occurrence.PLoS Comput Biol. 2016 Jul 29;12(7):e1005038. doi: 10.1371/journal.pcbi.1005038. eCollection 2016 Jul. PLoS Comput Biol. 2016. PMID: 27472895 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

