Postintegrative gene silencing within the Sleeping Beauty transposition system
- PMID: 17938204
- PMCID: PMC2169419
- DOI: 10.1128/MCB.00498-07
Postintegrative gene silencing within the Sleeping Beauty transposition system
Abstract
The Sleeping Beauty (SB) transposon represents an important vehicle for in vivo gene delivery because it can efficiently and stably integrate into mammalian genomes. In this report, we examined transposon expression in human cells using a novel nonselective fluorescence-activated cell sorter-based method and discovered that SB integrates approximately 20 times more frequently than previously reported within systems that were dependent on transgene expression and likely subject to postintegrative gene silencing. Over time, phenotypic analysis of clonal integrants demonstrated that SB undergoes additional postintegrative gene silencing, which varied based on the promoter used for transgene expression. Molecular and biochemical studies suggested that transposon silencing was influenced by DNA methylation and histone deacetylation because both 5-aza-2'-deoxycytidine and trichostatin A partially rescued transgene silencing in clonal cell lines. Collectively, these data reveal the existence of a multicomponent postintegrative gene silencing network that efficiently targets invading transposon sequences for transcriptional silencing in mammalian cells.
Figures
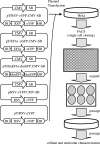

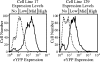
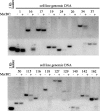
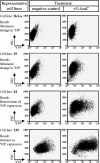
References
-
- Baus, J., L. Liu, A. D. Heggestad, S. Sanz, and B. S. Fletcher. 2005. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 12:1148-1156. - PubMed
-
- Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. - PubMed
-
- Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. - PubMed
-
- Christman, J. K. 2002. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21:5483-5495. - PubMed
-
- Dalle, B., J. E. Rubin, O. Alkan, T. Sukonnik, P. Pasceri, S. Yao, R. Pawliuk, P. Leboulch, and J. Ellis. 2005. eGFP reporter genes silence LCR β-globin transgene expression via CpG dinucleotides. Mol. Ther. 11:591-599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
