HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors
- PMID: 17938209
- PMCID: PMC2169410
- DOI: 10.1128/MCB.01724-07
HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors
Abstract
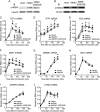
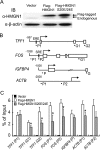
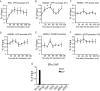
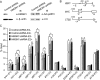
HMGN1, an abundant nucleosomal binding protein, can affect both the chromatin higher order structure and the modification of nucleosomal histones, but it alters the expression of only a subset of genes. We investigated specific gene targeting by HMGN1 in the context of estrogen induction of gene expression. Knockdown and overexpression experiments indicated that HMGN1 limits the induction of several estrogen-regulated genes, including TFF1 and FOS, which are induced by estrogen through entirely distinct mechanisms. HMGN1 specifically interacts with estrogen receptor alpha (ER alpha), both in vitro and in vivo. At the TFF1 promoter, estrogen increases HMGN1 association through recruitment by the ER alpha. HMGN1 S20E/S24E, although deficient in binding nucleosomal DNA, still interacts with ER alpha and, strikingly, still represses estrogen-driven activation of the TFF1 gene. On the FOS promoter, which lacks the ER alpha binding sites, constitutively bound serum response factor (SRF) mediates estrogen stimulation. HMGN1 also interacts specifically with SRF, but HMGN1 S20E/S24E does not. Consistent with the protein interactions, only wild-type HMGN1 significantly inhibits the estrogen-driven activation of the FOS gene. Mechanistically, the inhibition of estrogen induction of several ER alpha-associated genes, including TFF1, by HMGN1 correlates with decreased levels of acetylation of Lys9 on histone H3. Together, these findings indicate that HMGN1 regulates the expression of particular genes via specific protein-protein interactions with transcription factors at target gene regulatory regions.
Figures



Similar articles
-
Estrogen regulation of trefoil factor 1 expression by estrogen receptor alpha and Sp proteins.Exp Cell Res. 2005 Jan 1;302(1):96-107. doi: 10.1016/j.yexcr.2004.08.015. Exp Cell Res. 2005. PMID: 15541729
-
Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3.EMBO J. 2005 Sep 7;24(17):3038-48. doi: 10.1038/sj.emboj.7600768. Epub 2005 Aug 11. EMBO J. 2005. PMID: 16096646 Free PMC article.
-
Gene-specific patterns of coregulator requirements by estrogen receptor-α in breast cancer cells.Mol Endocrinol. 2012 Jun;26(6):955-66. doi: 10.1210/me.2012-1066. Epub 2012 Apr 27. Mol Endocrinol. 2012. PMID: 22543272 Free PMC article.
-
BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation.Mol Cell Biol. 2000 Oct;20(20):7541-9. doi: 10.1128/MCB.20.20.7541-7549.2000. Mol Cell Biol. 2000. PMID: 11003650 Free PMC article.
-
Chromosomal protein HMGN1 modulates histone H3 phosphorylation.Mol Cell. 2004 Aug 27;15(4):573-84. doi: 10.1016/j.molcel.2004.08.006. Mol Cell. 2004. PMID: 15327773
Cited by
-
KAT7/HMGN1 signaling epigenetically induces tyrosine phosphorylation-regulated kinase 1A expression to ameliorate insulin resistance in Alzheimer's disease.World J Psychiatry. 2024 Mar 19;14(3):445-455. doi: 10.5498/wjp.v14.i3.445. eCollection 2024 Mar 19. World J Psychiatry. 2024. PMID: 38617985 Free PMC article.
-
HMGN1 loss sensitizes lung cancer cells to chemotherapy.Sci Rep. 2024 May 6;14(1):10386. doi: 10.1038/s41598-024-60352-8. Sci Rep. 2024. PMID: 38710740 Free PMC article.
-
HMGN1 plays a significant role in CRLF2 driven Down Syndrome leukemia and provides a potential therapeutic target in this high-risk cohort.Oncogene. 2022 Feb;41(6):797-808. doi: 10.1038/s41388-021-02126-4. Epub 2021 Dec 2. Oncogene. 2022. PMID: 34857887
-
Pan-cancer Multi-omics Analysis Reveals HMGN1 as a Potential Prognostic and Immune Infiltration-associated Biomarker.Curr Med Chem. 2025;32(12):2440-2459. doi: 10.2174/0109298673268718231122103638. Curr Med Chem. 2025. PMID: 38310387 Free PMC article.
-
HMGNs, DNA repair and cancer.Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):80-5. doi: 10.1016/j.bbagrm.2009.10.007. Epub 2009 Dec 8. Biochim Biophys Acta. 2010. PMID: 20004154 Free PMC article. Review.
References
-
- Abercrombie, B. D., G. G. Kneale, C. Crane-Robinson, E. M. Bradbury, G. H. Goodwin, J. M. Walker, and E. W. Johns. 1978. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur. J. Biochem. 84:173-177. - PubMed
-
- Amano, T., K. Leu, K. Yoshizato, and Y. B. Shi. 2002. Thyroid hormone regulation of a transcriptional coactivator in Xenopus laevis: implication for a role in postembryonic tissue remodeling. Dev. Dyn. 223:526-535. - PubMed
-
- Augereau, P., F. Miralles, V. Cavailles, C. Gaudelet, M. Parker, and H. Rochefort. 1994. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol. Endocrinol. 8:693-703. - PubMed
-
- Begum, N., J. M. Pash, and J. S. Bhorjee. 1990. Expression and synthesis of high mobility group chromosomal proteins in different rat skeletal cell lines during myogenesis. J. Biol. Chem. 265:11936-11941. - PubMed
-
- Bertrand-Mercat, P., and J. R. Pasqualini. 1991. Antagonistic effect of the antiestrogen 4-hydroxytamoxifen on estradiol-stimulated acetylation of nuclear high mobility group (HMG) proteins in the uterus of newborn guinea pigs. Life Sci. 48:2081-2087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
