Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells
- PMID: 17938240
- PMCID: PMC2000320
- DOI: 10.1101/gad.1588207
Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells
Abstract
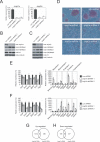
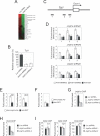
Embryonic stem (ES) cells are pluripotent cells with the ability to self-renew indefinitely. These unique properties are controlled by genetic factors and chromatin structure. The exit from the self-renewing state is accompanied by changes in epigenetic chromatin modifications such as an induction in the silencing-associated histone H3 Lys 9 dimethylation and trimethylation (H3K9Me2/Me3) marks. Here, we show that the H3K9Me2 and H3K9Me3 demethylase genes, Jmjd1a and Jmjd2c, are positively regulated by the ES cell transcription factor Oct4. Interestingly, Jmjd1a or Jmjd2c depletion leads to ES cell differentiation, which is accompanied by a reduction in the expression of ES cell-specific genes and an induction of lineage marker genes. Jmjd1a demethylates H3K9Me2 at the promoter regions of Tcl1, Tcfcp2l1, and Zfp57 and positively regulates the expression of these pluripotency-associated genes. Jmjd2c acts as a positive regulator for Nanog, which encodes for a key transcription factor for self-renewal in ES cells. We further demonstrate that Jmjd2c is required to reverse the H3K9Me3 marks at the Nanog promoter region and consequently prevents transcriptional repressors HP1 and KAP1 from binding. Our results connect the ES cell transcription circuitry to chromatin modulation through H3K9 demethylation in pluripotent cells.
Figures





Comment in
-
Open conformation chromatin and pluripotency.Genes Dev. 2007 Nov 1;21(21):2671-6. doi: 10.1101/gad.1615707. Genes Dev. 2007. PMID: 17974911 Review. No abstract available.
References
-
- Avilion A.A., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Nicolis S.K., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Pevny L.H., Perez L., Vivian N., Lovell-Badge R., Perez L., Vivian N., Lovell-Badge R., Vivian N., Lovell-Badge R., Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes & Dev. 2003;17:126–140. - PMC - PubMed
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Casanova M., Warnes G., Merkenschlager M., Warnes G., Merkenschlager M., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Zegerman P., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Partridge J.F., Miska E.A., Allshire R.C., Kouzarides T., Miska E.A., Allshire R.C., Kouzarides T., Allshire R.C., Kouzarides T., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Fry B., Meissner A., Wernig M., Plath K., Meissner A., Wernig M., Plath K., Wernig M., Plath K., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
-
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
