Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes
- PMID: 17938242
- PMCID: PMC2000323
- DOI: 10.1101/gad.1569307
Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes
Abstract
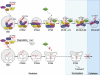
More than 170 proteins are necessary for assembly of ribosomes in eukaryotes. However, cofactors that function with each of these proteins, substrates on which they act, and the precise functions of assembly factors--e.g., recruiting other molecules into preribosomes or triggering structural rearrangements of pre-rRNPs--remain mostly unknown. Here we investigated the recruitment of two ribosomal proteins and 5S ribosomal RNA (rRNA) into nascent ribosomes. We identified a ribonucleoprotein neighborhood in preribosomes that contains two yeast ribosome assembly factors, Rpf2 and Rrs1, two ribosomal proteins, rpL5 and rpL11, and 5S rRNA. Interactions between each of these four proteins have been confirmed by binding assays in vitro. These molecules assemble into 90S preribosomal particles containing 35S rRNA precursor (pre-rRNA). Rpf2 and Rrs1 are required for recruiting rpL5, rpL11, and 5S rRNA into preribosomes. In the absence of association of these molecules with pre-rRNPs, processing of 27SB pre-rRNA is blocked. Consequently, the abortive 66S pre-rRNPs are prematurely released from the nucleolus to the nucleoplasm, and cannot be exported to the cytoplasm.
Figures






References
-
- Andrew C., Hopper A.K., Hall B.D., Hopper A.K., Hall B.D., Hall B.D. A yeast mutant defective in the processing of 27S r-RNA precursor. Mol. Gen. Genet. 1976;144:29–37. - PubMed
-
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Moore D.D., Seidman J.G., Smith J.A., Struhl K., Seidman J.G., Smith J.A., Struhl K., Smith J.A., Struhl K., Struhl K. Current protocols in molecular biology. John Wiley & Sons, Inc.; New York: 1994.
-
- Ballesta J.P., Remacha M., Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog. Nucleic Acids Res. Mol. Biol. 1996;55:157–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
