Plasma membrane domain organization regulates EGFR signaling in tumor cells
- PMID: 17938246
- PMCID: PMC2064769
- DOI: 10.1083/jcb.200611106
Plasma membrane domain organization regulates EGFR signaling in tumor cells
Abstract
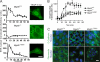
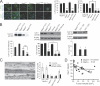
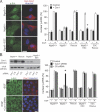
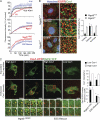
Macromolecular complexes exhibit reduced diffusion in biological membranes; however, the physiological consequences of this characteristic of plasma membrane domain organization remain elusive. We report that competition between the galectin lattice and oligomerized caveolin-1 microdomains for epidermal growth factor (EGF) receptor (EGFR) recruitment regulates EGFR signaling in tumor cells. In mammary tumor cells deficient for Golgi beta1,6N-acetylglucosaminyltransferase V (Mgat5), a reduction in EGFR binding to the galectin lattice allows an increased association with stable caveolin-1 cell surface microdomains that suppresses EGFR signaling. Depletion of caveolin-1 enhances EGFR diffusion, responsiveness to EGF, and relieves Mgat5 deficiency-imposed restrictions on tumor cell growth. In Mgat5(+/+) tumor cells, EGFR association with the galectin lattice reduces first-order EGFR diffusion rates and promotes receptor interaction with the actin cytoskeleton. Importantly, EGFR association with the lattice opposes sequestration by caveolin-1, overriding its negative regulation of EGFR diffusion and signaling. Therefore, caveolin-1 is a conditional tumor suppressor whose loss is advantageous when beta1,6GlcNAc-branched N-glycans are below a threshold for optimal galectin lattice formation.
Figures




References
-
- Ahmad, N., H.J. Gabius, S. Andre, H. Kaltner, S. Sabesan, R. Roy, B. Liu, F. Macaluso, and C.F. Brewer. 2004. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J. Biol. Chem. 279:10841–10847. - PubMed
-
- Breuza, L., S. Corby, J.P. Arsanto, M.H. Delgrossi, P. Scheiffele, and A. Le Bivic. 2002. The scaffolding domain of caveolin 2 is responsible for its Golgi localization in Caco-2 cells. J. Cell Sci. 115:4457–4467. - PubMed
-
- Brewer, C.F., M.C. Miceli, and L.G. Baum. 2002. Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 12:616–623. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

