Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase
- PMID: 17938735
- PMCID: PMC2022656
- DOI: 10.1289/ehp.10207
Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase
Abstract
Background: Aberrant DNA methylation is common in carcinogenesis. The typical pattern appears to involve reduced expression of maintenance DNA methyltransferase, DNMT1, inducing genomic hypomethylation, whereas increased expression of de novo DNMT3a or 3b causes gene-specific hypermethylation.
Objectives: During cadmium-induced malignant transformation, an unusual pattern of genomic hypermethylation occurred that we studied to provide insight into the roles of specific DNMTs in oncogenesis.
Methods: Gene expression and DNA methylation were assessed in control and chronic cadmium-transformed prostate epithelial cells (CTPE) using reverse transcription-polymerase chain reaction (RT-PCR), Western blot analysis, methylation-specific PCR, and methyl acceptance assay.
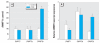
Results: During the 10-weeks of cadmium exposure that induced malignant transformation, progressive increases in generalized DNMT enzymatic activity occurred that were associated with over-expression of DNMT3b without changes in DNMT1 expression. Increased DNMT3b expression preceded increased DNMT enzymatic activity. Procainamide, a specific DNMT1 inhibitor, reversed cadmium-induced genomic DNA hypermethylation. Reduced expression of the tumor suppressor genes, RASSF1A and p16, began about the time DNMT3b overexpression first occurred and progressively decreased thereafter. RASSF1A and p16 promoter regions were heavily methylated in CTPE cells, indicating silencing by hypermethylation, while the DNA demethylating agent, 5-aza-2'-deoxycytidine, reversed this silencing. DNMT1 inhibition only modestly increased RASSF1A and p16 expression in CTPE cells and did not completely reverse silencing.
Conclusions: These data indicate that DNMT3b overexpression can result in generalized DNA hypermethylation and gene silencing but that DNMT1 is required to maintain these effects. The pattern of genomic DNA hypermethylation together with up-regulation of DNMT3b may provide a unique set of biomarkers to specifically identify cadmium-induced human prostate cancers.
Keywords: DNA methylation; DNMT3b; RASSF1A; cadmium; carcinogenesis; p16; prostate.
Figures




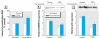
Similar articles
-
Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium.Toxicol Sci. 2012 Feb;125(2):412-7. doi: 10.1093/toxsci/kfr320. Epub 2011 Nov 23. Toxicol Sci. 2012. PMID: 22112500
-
Aberrant methylation accounts for cell adhesion-related gene silencing during 3-methylcholanthrene and diethylnitrosamine induced multistep rat lung carcinogenesis associated with overexpression of DNA methyltransferases 1 and 3a.Toxicol Appl Pharmacol. 2011 Feb 15;251(1):70-8. doi: 10.1016/j.taap.2010.12.002. Epub 2010 Dec 14. Toxicol Appl Pharmacol. 2011. PMID: 21163286
-
Long-term cadmium exposure leads to the enhancement of lymphocyte proliferation via down-regulating p16 by DNA hypermethylation.Mutat Res. 2013 Oct 9;757(2):125-31. doi: 10.1016/j.mrgentox.2013.07.007. Epub 2013 Aug 12. Mutat Res. 2013. PMID: 23948183
-
DNA methyltransferase-1 inhibitors as epigenetic therapy for cancer.Curr Cancer Drug Targets. 2013 May;13(4):379-99. doi: 10.2174/15680096113139990077. Curr Cancer Drug Targets. 2013. PMID: 23517596 Review.
-
Tissue-specific roles of de novo DNA methyltransferases.Epigenetics Chromatin. 2025 Jan 17;18(1):5. doi: 10.1186/s13072-024-00566-2. Epigenetics Chromatin. 2025. PMID: 39819598 Free PMC article. Review.
Cited by
-
Low-level environmental cadmium exposure is associated with DNA hypomethylation in Argentinean women.Environ Health Perspect. 2012 Jun;120(6):879-84. doi: 10.1289/ehp.1104600. Epub 2012 Mar 1. Environ Health Perspect. 2012. PMID: 22382075 Free PMC article.
-
Epigenetic Contributions to the Relationship between Cancer and Dietary Intake of Nutrients, Bioactive Food Components, and Environmental Toxicants.Front Genet. 2012 Jan 9;2:91. doi: 10.3389/fgene.2011.00091. eCollection 2011. Front Genet. 2012. PMID: 22303385 Free PMC article.
-
Nc886 is epigenetically repressed in prostate cancer and acts as a tumor suppressor through the inhibition of cell growth.BMC Cancer. 2018 Feb 2;18(1):127. doi: 10.1186/s12885-018-4049-7. BMC Cancer. 2018. PMID: 29394925 Free PMC article.
-
In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning.J Exp Bot. 2012 Jan;63(2):695-709. doi: 10.1093/jxb/err313. Epub 2011 Nov 4. J Exp Bot. 2012. PMID: 22058406 Free PMC article.
-
Environmental pollution and DNA methylation: carcinogenesis, clinical significance, and practical applications.Front Med. 2015 Sep;9(3):261-74. doi: 10.1007/s11684-015-0406-y. Epub 2015 Aug 19. Front Med. 2015. PMID: 26290283 Review.
References
-
- Achanzar WE, Diwan BA, Liu J, Quader ST, Webber MM, Waalkes MP. Cadmium-induced malignant transformation of human prostate epithelial cells. Cancer Res. 2001;61(2):455–458. - PubMed
-
- Auerkari EI. Methylation of tumor suppressor genes p16(INK4a), p27(Kip1) and E-cadherin in carcinogenesis. Oral Oncology. 2006;42(1):5–13. - PubMed
-
- Balaghi M, Wagner C. DNA methylation in folate deficiency: use of CpG methylase. Biochem Biophys Res Commun. 1993;93(3):1184–1190. - PubMed
-
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. - PubMed
-
- Baylin SB, Makos M, Wu JJ, Yen RW, de Butros A, Vertino P, et al. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991;3(10):383–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

