Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers
- PMID: 17940009
- PMCID: PMC2040403
- DOI: 10.1073/pnas.0708210104
Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers
Abstract
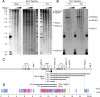
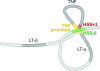
Here we provide a mechanism for specific, efficient transcription of the TNF gene and, potentially, other genes residing within multigene loci. We identify and characterize highly conserved noncoding elements flanking the TNF gene, which undergo activation-dependent intrachromosomal interactions. These elements, hypersensitive site (HSS)-9 and HSS+3 (9 kb upstream and 3 kb downstream of the TNF gene, respectively), contain DNase I hypersensitive sites in naive, T helper 1, and T helper 2 primary T cells. Both HSS-9 and HSS+3 inducibly associate with acetylated histones, indicative of chromatin remodeling, bind the transcription factor nuclear factor of activated T cells (NFAT)p in vitro and in vivo, and function as enhancers of NFAT-dependent transactivation mediated by the TNF promoter. Using the chromosome conformation capture assay, we demonstrate that upon T cell activation intrachromosomal looping occurs in the TNF locus. HSS-9 and HSS+3 each associate with the TNF promoter and with each other, circularizing the TNF gene and bringing NFAT-containing nucleoprotein complexes into close proximity. TNF gene regulation thus reveals a mode of intrachromosomal interaction that combines a looped gene topology with interactions between enhancers and a gene promoter.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation.Adv Immunol. 2013;118:37-128. doi: 10.1016/B978-0-12-407708-9.00002-9. Adv Immunol. 2013. PMID: 23683942 Free PMC article. Review.
-
NFAT is well placed to direct both enhancer looping and domain-wide models of enhancer function.Sci Signal. 2008 Apr 1;1(13):pe15. doi: 10.1126/stke.113pe15. Sci Signal. 2008. PMID: 18385038
-
Identification of a Distal Locus Enhancer Element That Controls Cell Type-Specific TNF and LTA Gene Expression in Human T Cells.J Immunol. 2020 Nov 1;205(9):2479-2488. doi: 10.4049/jimmunol.1901311. Epub 2020 Sep 25. J Immunol. 2020. PMID: 32978279 Free PMC article.
-
Chromatin profiling across the human tumour necrosis factor gene locus reveals a complex, cell type-specific landscape with novel regulatory elements.Nucleic Acids Res. 2008 Sep;36(15):4845-62. doi: 10.1093/nar/gkn444. Epub 2008 Jul 24. Nucleic Acids Res. 2008. PMID: 18653526 Free PMC article.
-
Mechanisms of transcriptional regulation of the human IL-3/GM-CSF locus by inducible tissue-specific promoters and enhancers.Crit Rev Immunol. 2004;24(6):385-408. doi: 10.1615/critrevimmunol.v24.i6.10. Crit Rev Immunol. 2004. PMID: 15777160 Review.
Cited by
-
Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation.Adv Immunol. 2013;118:37-128. doi: 10.1016/B978-0-12-407708-9.00002-9. Adv Immunol. 2013. PMID: 23683942 Free PMC article. Review.
-
An epigenetic chromatin remodeling role for NFATc1 in transcriptional regulation of growth and survival genes in diffuse large B-cell lymphomas.Blood. 2010 Nov 11;116(19):3899-906. doi: 10.1182/blood-2009-12-257378. Epub 2010 Jul 27. Blood. 2010. PMID: 20664054 Free PMC article.
-
Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9.Nat Commun. 2021 Sep 29;12(1):5711. doi: 10.1038/s41467-021-25991-9. Nat Commun. 2021. PMID: 34588447 Free PMC article.
-
Higher-Order Chromatin Regulation of Inflammatory Gene Expression.Mediators Inflamm. 2017;2017:7848591. doi: 10.1155/2017/7848591. Epub 2017 Apr 9. Mediators Inflamm. 2017. PMID: 28490839 Free PMC article. Review.
-
IL-10 inhibits transcription elongation of the human TNF gene in primary macrophages.J Exp Med. 2010 Sep 27;207(10):2081-8. doi: 10.1084/jem.20100414. Epub 2010 Aug 30. J Exp Med. 2010. PMID: 20805562 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

