Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus
- PMID: 17940010
- PMCID: PMC2040464
- DOI: 10.1073/pnas.0708209104
Antagonistic pleiotropic effects reduce the potential adaptive value of the FRIGIDA locus
Abstract
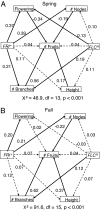
Although the occurrence of epistasis and pleiotropy is widely accepted at the molecular level, its effect on the adaptive value of fitness-related genes is rarely investigated in plants. Knowledge of these features of a gene is critical to understand the molecular basis of adaptive evolution. Here we investigate the importance of pleiotropy and epistasis in determining the adaptive value of a candidate gene using the gene FRI (FRIGIDA), which is thought to be the major gene controlling flowering time variation in Arabidopsis thaliana. The effect of FRI on flowering time was analyzed in an outbred population created by randomly mating 19 natural accessions of A. thaliana. This unique population allows the estimation of FRI effects independent of any linkage association with other loci due to demographic processes or to coadapted genes. It also allows for the estimation of pleiotropic effects of FRI on fitness and inflorescence architecture. We found that FRI explains less variation in flowering time than previously observed among natural accessions, and interacts epistatically with the FLC locus. Although early flowering plants produce more fruits under spring conditions, and nonfunctional alleles of FRI were associated with early flowering, variation at FRI was not associated with fitness. We show that nonfunctional FRI alleles have negative pleiotropic effects on fitness by reducing the numbers of nodes and branches on the inflorescence. We propose that these antagonistic pleiotropic effects reduce the adaptive value of FRI, and helps explain the maintenance of alternative life history strategies across natural populations of A. thaliana.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Standing genetic variation in FRIGIDA mediates experimental evolution of flowering time in Arabidopsis.Mol Ecol. 2009 May;18(9):2039-49. doi: 10.1111/j.1365-294X.2009.04145.x. Epub 2009 Mar 20. Mol Ecol. 2009. PMID: 19317844
-
FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions.Genetics. 2005 Jul;170(3):1197-207. doi: 10.1534/genetics.104.036533. Epub 2005 May 23. Genetics. 2005. PMID: 15911588 Free PMC article.
-
Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis.Plant Physiol. 2005 Jun;138(2):1163-73. doi: 10.1104/pp.105.061309. Epub 2005 May 20. Plant Physiol. 2005. PMID: 15908596 Free PMC article.
-
Role of chromatin modification in flowering-time control.Trends Plant Sci. 2005 Jan;10(1):30-5. doi: 10.1016/j.tplants.2004.11.003. Trends Plant Sci. 2005. PMID: 15642521 Review.
-
Pleiotropy in developmental regulation by flowering-pathway genes: is it an evolutionary constraint?New Phytol. 2019 Oct;224(1):55-70. doi: 10.1111/nph.15901. Epub 2019 Jun 18. New Phytol. 2019. PMID: 31074008 Review.
Cited by
-
Life-history QTLS and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis.Evolution. 2011 Mar;65(3):771-87. doi: 10.1111/j.1558-5646.2010.01175.x. Epub 2010 Nov 17. Evolution. 2011. PMID: 21083662 Free PMC article.
-
Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana.Genes Dev. 2014 Aug 1;28(15):1635-40. doi: 10.1101/gad.245993.114. Epub 2014 Jul 17. Genes Dev. 2014. PMID: 25035417 Free PMC article.
-
Adaptation via pleiotropy and linkage: Association mapping reveals a complex genetic architecture within the stickleback Eda locus.Evol Lett. 2020 May 27;4(4):282-301. doi: 10.1002/evl3.175. eCollection 2020 Aug. Evol Lett. 2020. PMID: 32774879 Free PMC article.
-
Flowering time in maize: linkage and epistasis at a major effect locus.Genetics. 2012 Apr;190(4):1547-62. doi: 10.1534/genetics.111.136903. Epub 2012 Jan 31. Genetics. 2012. PMID: 22298708 Free PMC article.
-
The evolution of metabolism: How to test evolutionary hypotheses at the genomic level.Comput Struct Biotechnol J. 2020 Feb 20;18:482-500. doi: 10.1016/j.csbj.2020.02.009. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32180906 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

