Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing
- PMID: 17940139
- PMCID: PMC2080592
- DOI: 10.1261/rna.626207
Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing
Abstract
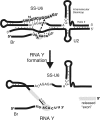
Splicing of introns from mRNA precursors is a two-step reaction performed by the spliceosome, an immense cellular machine consisting of over 200 different proteins and five small RNAs (snRNAs). We previously demonstrated that fragments of two of these RNAs, U6 and U2, can catalyze by themselves a splicing-related reaction, involving one of the two substrates of the first step of splicing, the branch site substrate. Here we show that these same RNAs can catalyze a reaction between RNA sequences that resemble the 5' splice site and the branch site, the two reactants of the first step of splicing. The reaction is dependent on the sequence of the 5' splice site consensus sequence and the catalytically essential domains of U6, and thus it resembles the authentic splicing reaction. Our results demonstrate the ability of protein-free snRNAs to recognize the sequences involved in the first splicing step and to perform splicing-related catalysis between these two pre-mRNA-like substrates.
Figures
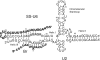


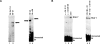
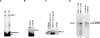

Similar articles
-
Splicing-related catalysis by protein-free snRNAs.Nature. 2001 Oct 18;413(6857):701-7. doi: 10.1038/35099500. Nature. 2001. PMID: 11607023
-
The role of snRNAs in spliceosomal catalysis.Prog Mol Biol Transl Sci. 2013;120:195-228. doi: 10.1016/B978-0-12-381286-5.00006-8. Prog Mol Biol Transl Sci. 2013. PMID: 24156945
-
Splicing of an intervening sequence by protein-free human snRNAs.RNA Biol. 2011 May-Jun;8(3):372-7. doi: 10.4161/rna.8.3.15386. Epub 2011 May 1. RNA Biol. 2011. PMID: 21445000 Free PMC article.
-
Role of the snRNAs in spliceosomal active site.RNA Biol. 2010 May-Jun;7(3):345-53. doi: 10.4161/rna.7.3.12089. Epub 2010 May 16. RNA Biol. 2010. PMID: 20458185 Review.
-
The spliceosome: a ribozyme at heart?Biol Chem. 2007 Jul;388(7):693-7. doi: 10.1515/BC.2007.080. Biol Chem. 2007. PMID: 17570821 Review.
Cited by
-
Protein-free small nuclear RNAs catalyze a two-step splicing reaction.Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):11901-6. doi: 10.1073/pnas.0902020106. Epub 2009 Jun 22. Proc Natl Acad Sci U S A. 2009. PMID: 19549866 Free PMC article.
-
Intron Retention and Alzheimer's Disease (AD): A Review of Regulation Genes Implicated in AD.Genes (Basel). 2025 Jun 30;16(7):782. doi: 10.3390/genes16070782. Genes (Basel). 2025. PMID: 40725435 Free PMC article. Review.
-
Role of the central junction in folding topology of the protein-free human U2-U6 snRNA complex.RNA. 2020 Jul;26(7):836-850. doi: 10.1261/rna.073379.119. Epub 2020 Mar 27. RNA. 2020. PMID: 32220895 Free PMC article.
-
Comparison of Alternative Splicing Junction Detection Tools Using RNA-Seq Data.Curr Genomics. 2017 Jun;18(3):268-277. doi: 10.2174/1389202918666170215125048. Curr Genomics. 2017. PMID: 28659722 Free PMC article. Review.
-
Group II intron lariat: Structural insights into the spliceosome.RNA Biol. 2015;12(9):913-7. doi: 10.1080/15476286.2015.1066956. RNA Biol. 2015. PMID: 26121424 Free PMC article. Review.
References
-
- Brow, D.A. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 2002;36:333–360. - PubMed
-
- Burke, J.M., Butcher, S.E., Sargueil, B. Structural analysis and modification of the hairpin ribozyme. In: Eckstein F., Lilley D.M.J., editors. Nucleic acids and molecular biology. Springer; Berlin, Germany: 1996. pp. 129–143.
-
- Collins, C.A., Guthrie, C. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 2000;7:850–854. - PubMed
-
- Cowan, J.A. Metallobiochemistry of RNA. Co(NH3)6 3+ as a probe for Mg2+(aq) binding sites. J. Inorg. Biochem. 1993;49:171–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
