Selective incorporation of polyanionic molecules into hamster prions
- PMID: 17940287
- PMCID: PMC3091164
- DOI: 10.1074/jbc.M704447200
Selective incorporation of polyanionic molecules into hamster prions
Abstract
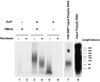
The central pathogenic event of prion disease is the conformational conversion of a host protein, PrPC, into a pathogenic isoform, PrPSc. We previously showed that the protein misfolding cyclic amplification (PMCA) technique can be used to form infectious prion molecules de novo from purified native PrPC molecules in an autocatalytic process requiring accessory polyanions (Deleault, N. R., Harris, B. T., Rees, J. R., and Supattapone, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104, 9741-9746). Here we investigated the molecular mechanism by which polyanionic molecules facilitate infectious prion formation in vitro. Ina PMCA reaction lacking PrPSc template seed, synthetic polyA RNA molecules induce hamster HaPrPC to adopt a protease-sensitive, detergent-insoluble conformation reactive against antibodies specific for PrPSc. During PMCA, labeled nucleic acids form nuclease-resistant complexes with HaPrP molecules. Strikingly, purified HaPrPC molecules subjected to PMCA selectively incorporate an approximately 1-2.5-kb subset of [32P]polyA RNA molecules from a heterogeneous mixture ranging in size from approximately 0.1 to >6 kb. Neuropathological analysis of scrapie-infected hamsters using the fluorescent dye acridine orange revealed that RNA molecules co-localize with large extracellular HaPrP aggregates. These findings suggest that polyanionic molecules such as RNA may become selectively incorporated into stable complexes with PrP molecules during the formation of native hamster prions.
Figures




References
-
- Glatzel M, Stoeck K, Seeger H, Luhrs T, Aguzzi A. Arch. Neurol. 2005;62:545–552. - PubMed
-
- Prusiner SB. Science. 1982;216:136–144. - PubMed
-
- Telling GC, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen FE, DeArmond SJ, Prusiner SB. Cell. 1995;83:79–90. - PubMed
-
- Saborio GP, Soto C, Kascsak RJ, Levy E, Kascsak R, Harris DA, Frangione B. Biochem. Biophys. Res. Commun. 1999;258:470–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

