Network-based classification of breast cancer metastasis
- PMID: 17940530
- PMCID: PMC2063581
- DOI: 10.1038/msb4100180
Network-based classification of breast cancer metastasis
Abstract
Mapping the pathways that give rise to metastasis is one of the key challenges of breast cancer research. Recently, several large-scale studies have shed light on this problem through analysis of gene expression profiles to identify markers correlated with metastasis. Here, we apply a protein-network-based approach that identifies markers not as individual genes but as subnetworks extracted from protein interaction databases. The resulting subnetworks provide novel hypotheses for pathways involved in tumor progression. Although genes with known breast cancer mutations are typically not detected through analysis of differential expression, they play a central role in the protein network by interconnecting many differentially expressed genes. We find that the subnetwork markers are more reproducible than individual marker genes selected without network information, and that they achieve higher accuracy in the classification of metastatic versus non-metastatic tumors.
Figures
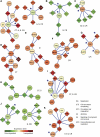
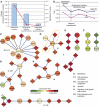
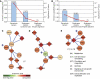
Comment in
-
Protein subnetwork markers improve prediction of cancer outcome.Mol Syst Biol. 2007;3:141. doi: 10.1038/msb4100183. Epub 2007 Oct 16. Mol Syst Biol. 2007. PMID: 17940531 Free PMC article. No abstract available.
References
-
- Agresti A (1990) Categorical Data Analysis. New York: Wiley
-
- Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D'Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H et al.. (2005) The biomolecular interaction network database and related tools 2005 update. Nucleic Acids Res 33: D418–D424 - PMC - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC et al.. (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403: 503–511 - PubMed
-
- Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH (2004) The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 3: 772–775 - PubMed
-
- Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z (2000) Tissue classification with gene expression profiles. J Comput Biol 7: 559–583 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

