Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose
- PMID: 17941826
- PMCID: PMC4379490
- DOI: 10.1042/BJ20071044
Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose
Abstract
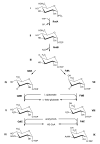
Derivatives of 3-amino-3,6-dideoxyhexoses are widespread in Nature. They are part of the repeating units of lipopolysaccharide O-antigens, of the glycan moiety of S-layer (bacterial cell surface layer) glycoproteins and also of many antibiotics. In the present study, we focused on the elucidation of the biosynthesis pathway of dTDP-alpha-D-Quip3NAc (dTDP-3-acetamido-3,6-dideoxy-alpha-D-glucose) from the Gram-positive, anaerobic, thermophilic organism Thermoanaerobacterium thermosaccharolyticum E207-71, which carries Quip3NAc in its S-layer glycan. The biosynthesis of dTDP-alpha-D-Quip3NAc involves five enzymes, namely a transferase, a dehydratase, an isomerase, a transaminase and a transacetylase, and follows a pathway similar to that of dTDP-alpha-D-Fucp3NAc (dTDP-3-acetamido-3,6-dideoxy-alpha-D-galactose) biosynthesis in Aneurinibacillus thermoaerophilus L420-91(T). The ORFs (open reading frames) of interest were cloned, overexpressed in Escherichia coli and purified. To elucidate the enzymatic cascade, the different products were purified by HPLC and characterized by NMR spectroscopy. The initiating reactions catalysed by the glucose-1-phosphate thymidylyltransferase RmlA and the dTDP-D-glucose-4,6-dehydratase RmlB are well established. The subsequent isomerase was shown to be capable of forming a dTDP-3-oxo-6-deoxy-D-glucose intermediate from the RmlB product dTDP-4-oxo-6-deoxy-D-glucose, whereas the isomerase involved in the dTDP-alpha-D-Fucp3NAc pathway synthesizes dTDP-3-oxo-6-deoxy-D-galactose. The subsequent reaction steps of either pathway involve a transaminase and a transacetylase, leading to the specific production of nucleotide-activated 3-acetamido-3,6-dideoxy-alpha-D-glucose and 3-acetamido-3,6-dideoxy-alpha-D-galactose respectively. Sequence comparison of the ORFs responsible for the biosynthesis of dTDP-alpha-D-Quip3NAc revealed homologues in Gram-negative as well as in antibiotic-producing Gram-positive bacteria. There is strong evidence that the elucidated biosynthesis pathway may also be valid for LPS (lipopolysaccharide) O-antigen structures and antibiotic precursors.
Figures


References
-
- Messner P, Schäffer C. Prokaryotic glycoproteins. In: Herz W, Falk H, Kirby GW, editors. Progress in the Chemistry of Organic Natural Products. Volume 85. Springer, Wien, Austria, and New York: 2003. pp. 51–124. - PubMed
-
- Keenleyside WJ, Whitfield C. Genetics and biosynthesis of lipopolysaccharides. In: Brade H, Opal SN, Vogel SN, Morrison DC, editors. Endotoxins in Health and Disease. Marcel Dekker; New York: 1999. pp. 331–358.
-
- Graninger M, Kneidinger B, Bruno K, Scheberl A, Messner P. Homologs of the Rml enzymes from Salmonella enterica are responsible for dTDP-β-l-rhamnose biosynthesis in the gram-positive thermophile Aneurinibacillus thermoaerophilus DSM 10155. Appl. Environ. Microbiol. 2002;68:3708–3715. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

