Structural analysis of kinetic folding intermediates for a TIM barrel protein, indole-3-glycerol phosphate synthase, by hydrogen exchange mass spectrometry and Gō model simulation
- PMID: 17942114
- PMCID: PMC2735044
- DOI: 10.1016/j.jmb.2007.09.024
Structural analysis of kinetic folding intermediates for a TIM barrel protein, indole-3-glycerol phosphate synthase, by hydrogen exchange mass spectrometry and Gō model simulation
Abstract
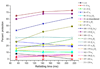
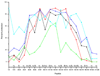
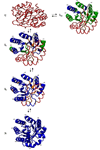
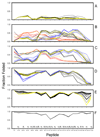
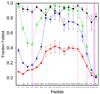
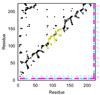
The structures of partially folded states appearing during the folding of a (betaalpha)(8) TIM barrel protein, the indole-3-glycerol phosphate synthase from Sulfolobus solfataricus (sIGPS), was assessed by hydrogen exchange mass spectrometry (HX-MS) and Gō model simulations. HX-MS analysis of the peptic peptides derived from the pulse-labeled product of the sub-millisecond folding reaction from the urea-denatured state revealed strong protection in the (betaalpha)(4) region, modest protection in the neighboring (betaalpha)(1-3) and (betaalpha)(5)beta(6) segments and no significant protection in the remaining N and C-terminal segments. These results demonstrate that this species is not a collapsed form of the unfolded state under native-favoring conditions nor is it the native state formed via fast-track folding. However, the striking contrast of these results with the strong protection observed in the (betaalpha)(2-5)beta(6) region after 5 s of folding demonstrates that these species represent kinetically distinct folding intermediates that are not identical as previously thought. A re-examination of the kinetic folding mechanism by chevron analysis of fluorescence data confirmed distinct roles for these two species: the burst-phase intermediate is predicted to be a misfolded, off-pathway intermediate, while the subsequent 5 s intermediate corresponds to an on-pathway equilibrium intermediate. Comparison with the predictions using a C(alpha) Gō model simulation of the kinetic folding reaction for sIGPS shows good agreement with the core of the structure offering protection against exchange in the on-pathway intermediate(s). Because the native-centric Gō model simulations do not explicitly include sequence-specific information, the simulation results support the hypothesis that the topology of TIM barrel proteins is a primary determinant of the folding free energy surface for the productive folding reaction. The early misfolding reaction must involve aspects of non-native structure not detected by the Gō model simulation.
Figures




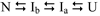
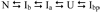
References
-
- Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–R91. - PubMed
-
- Ballew RM, Sabelko J, Gruebele M. Observation of distinct nanosecond and microsecond protein folding events. Nat Struct Biol. 1996;3:923–926. - PubMed
-
- Forsyth WR, Matthews CR. Folding Mechanism of Indole-3-glycerol Phosphate Synthase from Sulfolobus solfataricus: A Test of the Conservation of Folding Mechanisms Hypothesis in (βα)8 Barrels. J Mol Biol. 2002;320:1119–1133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

