Ohno's dilemma: evolution of new genes under continuous selection
- PMID: 17942681
- PMCID: PMC2040452
- DOI: 10.1073/pnas.0707158104
Ohno's dilemma: evolution of new genes under continuous selection
Abstract
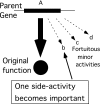
New genes with novel functions arise by duplication and divergence, but the process poses a problem. After duplication, an extra gene copy must rise to sufficiently high frequency in the population and remain free of common inactivating lesions long enough to acquire the rare mutations that provide a new selectable function. Maintaining a duplicated gene by selection for the original function would restrict the freedom to diverge. (We refer to this problem as Ohno's dilemma). A model is described by which selection continuously favors both maintenance of the duplicate copy and divergence of that copy from the parent gene. Before duplication, the original gene has a trace side activity (the innovation) in addition to its original function. When an altered ecological niche makes the minor innovation valuable, selection favors increases in its level (the amplification), which is most frequently conferred by increased dosage of the parent gene. Selection for the amplified minor function maintains the extra copies and raises the frequency of the amplification in the population. The same selection favors mutational improvement of any of the extra copies, which are not constrained to maintain their original function (the divergence). The rate of mutations (per genome) that improve the new function is increased by the multiplicity of target copies within a genome. Improvement of some copies relaxes selection on others and allows their loss by mutation (becoming pseudogenes). Ultimately one of the extra copies is able to provide all of the new activity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

