Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability
- PMID: 17942694
- PMCID: PMC2040459
- DOI: 10.1073/pnas.0704008104
Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability
Abstract
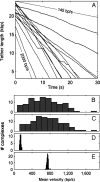
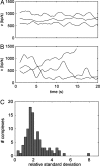
Terminase enzyme complexes, which facilitate ATP-driven DNA packaging in phages and in many eukaryotic viruses, constitute a wide and potentially diverse family of molecular motors about which little dynamic or mechanistic information is available. Here we report optical tweezers measurements of single DNA molecule packaging dynamics in phage T4, a large, tailed Escherichia coli virus that is an important model system in molecular biology. We show that a complex is formed between the empty prohead and the large terminase protein (gp17) that can capture and begin packaging a target DNA molecule within a few seconds, thus demonstrating a distinct viral assembly pathway. The motor generates forces >60 pN, similar to those measured with phage phi29, suggesting that high force generation is a common property of viral DNA packaging motors. However, the DNA translocation rate for T4 was strikingly higher than that for phi29, averaging approximately 700 bp/s and ranging up to approximately 2,000 bp/s, consistent with packaging by phage T4 of an enormous, 171-kb genome in <10 min during viral infection and implying high ATP turnover rates of >300 s(-1). The motor velocity decreased with applied load but averaged 320 bp/s at 45 pN, indicating very high power generation. Interestingly, the motor also exhibited large dynamic changes in velocity, suggesting that it can assume multiple active conformational states gearing different translocation rates. This capability, in addition to the reversible pausing and slipping capabilities that were observed, may allow phage T4 to coordinate DNA packaging with other ongoing processes, including viral DNA transcription, recombination, and repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4.Proc Natl Acad Sci U S A. 2012 Dec 4;109(49):20000-5. doi: 10.1073/pnas.1209214109. Epub 2012 Nov 19. Proc Natl Acad Sci U S A. 2012. PMID: 23169641 Free PMC article.
-
Single-Molecule Measurements of Motor-Driven Viral DNA Packaging in Bacteriophages Phi29, Lambda, and T4 with Optical Tweezers.Methods Mol Biol. 2018;1805:393-422. doi: 10.1007/978-1-4939-8556-2_20. Methods Mol Biol. 2018. PMID: 29971729
-
Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages.J Biol Chem. 2011 Feb 4;286(5):3944-56. doi: 10.1074/jbc.M110.196907. Epub 2010 Dec 2. J Biol Chem. 2011. PMID: 21127059 Free PMC article.
-
Old, new, and widely true: The bacteriophage T4 DNA packaging mechanism.Virology. 2015 May;479-480:650-6. doi: 10.1016/j.virol.2015.01.015. Epub 2015 Feb 27. Virology. 2015. PMID: 25728298 Free PMC article. Review.
-
DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism.Bioessays. 1995 Dec;17(12):1025-30. doi: 10.1002/bies.950171206. Bioessays. 1995. PMID: 8634063 Review.
Cited by
-
Single-molecule measurements of viral ssRNA packaging.RNA. 2017 Jan;23(1):119-129. doi: 10.1261/rna.057471.116. Epub 2016 Nov 1. RNA. 2017. PMID: 27803153 Free PMC article.
-
Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic acid motors.Methods Mol Biol. 2012;875:85-104. doi: 10.1007/978-1-61779-806-1_5. Methods Mol Biol. 2012. PMID: 22573437 Free PMC article.
-
The T4 TerL Prohead Packaging Motor Does Not Drive DNA Translocation by a Proposed Dehydration Mechanism.Viruses. 2020 May 9;12(5):522. doi: 10.3390/v12050522. Viruses. 2020. PMID: 32397493 Free PMC article.
-
Molecular architecture of tailed double-stranded DNA phages.Bacteriophage. 2014 Jan 1;4(1):e28281. doi: 10.4161/bact.28281. Epub 2014 Feb 21. Bacteriophage. 2014. PMID: 24616838 Free PMC article. Review.
-
Artificial bio-nanomachines based on protein needles derived from bacteriophage T4.Biophys Rev. 2018 Apr;10(2):641-658. doi: 10.1007/s12551-017-0336-9. Epub 2017 Nov 16. Biophys Rev. 2018. PMID: 29147941 Free PMC article. Review.
References
-
- Jardine PJ, Anderson D. In: The Bacteriophages. Calendar RL, editor. New York: Oxford Univ Press; 2006. pp. 49–65.
-
- Feiss M, Catalano CE. In: Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Catalano CE, editor. New York: Springer; 2005. pp. 5–39.
-
- Black LW. Annu Rev Microbiol. 1989;43:267–292. - PubMed
-
- Leiman PG, Chipman PR, Kostyuchenko VA, Mesyanzhinov VV, Rossmann MG. Cell. 2004;118:419–429. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

