A second generation human haplotype map of over 3.1 million SNPs
- PMID: 17943122
- PMCID: PMC2689609
- DOI: 10.1038/nature06258
A second generation human haplotype map of over 3.1 million SNPs
Abstract
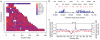
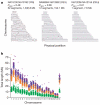
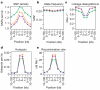
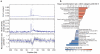
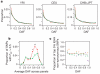
We describe the Phase II HapMap, which characterizes over 3.1 million human single nucleotide polymorphisms (SNPs) genotyped in 270 individuals from four geographically diverse populations and includes 25-35% of common SNP variation in the populations surveyed. The map is estimated to capture untyped common variation with an average maximum r2 of between 0.9 and 0.96 depending on population. We demonstrate that the current generation of commercial genome-wide genotyping products captures common Phase II SNPs with an average maximum r2 of up to 0.8 in African and up to 0.95 in non-African populations, and that potential gains in power in association studies can be obtained through imputation. These data also reveal novel aspects of the structure of linkage disequilibrium. We show that 10-30% of pairs of individuals within a population share at least one region of extended genetic identity arising from recent ancestry and that up to 1% of all common variants are untaggable, primarily because they lie within recombination hotspots. We show that recombination rates vary systematically around genes and between genes of different function. Finally, we demonstrate increased differentiation at non-synonymous, compared to synonymous, SNPs, resulting from systematic differences in the strength or efficacy of natural selection between populations.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

