Genome-wide detection and characterization of positive selection in human populations
- PMID: 17943131
- PMCID: PMC2687721
- DOI: 10.1038/nature06250
Genome-wide detection and characterization of positive selection in human populations
Abstract
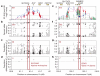
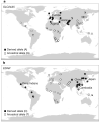
With the advent of dense maps of human genetic variation, it is now possible to detect positive natural selection across the human genome. Here we report an analysis of over 3 million polymorphisms from the International HapMap Project Phase 2 (HapMap2). We used 'long-range haplotype' methods, which were developed to identify alleles segregating in a population that have undergone recent selection, and we also developed new methods that are based on cross-population comparisons to discover alleles that have swept to near-fixation within a population. The analysis reveals more than 300 strong candidate regions. Focusing on the strongest 22 regions, we develop a heuristic for scrutinizing these regions to identify candidate targets of selection. In a complementary analysis, we identify 26 non-synonymous, coding, single nucleotide polymorphisms showing regional evidence of positive selection. Examination of these candidates highlights three cases in which two genes in a common biological process have apparently undergone positive selection in the same population:LARGE and DMD, both related to infection by the Lassa virus, in West Africa;SLC24A5 and SLC45A2, both involved in skin pigmentation, in Europe; and EDAR and EDA2R, both involved in development of hair follicles, in Asia.
Figures

References
-
- Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. - PubMed
-
- Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum. Mutat. 2005;25:278–284. - PubMed
-
- Lamason RL, et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

