From microscopes to microarrays: dissecting recurrent chromosomal rearrangements
- PMID: 17943194
- PMCID: PMC2858421
- DOI: 10.1038/nrg2136
From microscopes to microarrays: dissecting recurrent chromosomal rearrangements
Abstract
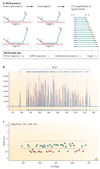
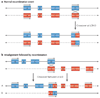
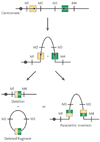
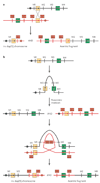
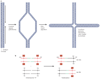
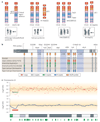
Submicroscopic chromosomal rearrangements that lead to copy-number changes have been shown to underlie distinctive and recognizable clinical phenotypes. The sensitivity to detect copy-number variation has escalated with the advent of array comparative genomic hybridization (CGH), including BAC and oligonucleotide-based platforms. Coupled with improved assemblies and annotation of genome sequence data, these technologies are facilitating the identification of new syndromes that are associated with submicroscopic genomic changes. Their characterization reveals the role of genome architecture in the aetiology of many clinical disorders. We review a group of genomic disorders that are mediated by segmental duplications, emphasizing the impact that high-throughput detection methods and the availability of the human genome sequence have had on their dissection and diagnosis.
Figures
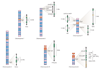
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

