Species-specific evolution of immune receptor tyrosine based activation motif-containing CEACAM1-related immune receptors in the dog
- PMID: 17945019
- PMCID: PMC2110893
- DOI: 10.1186/1471-2148-7-196
Species-specific evolution of immune receptor tyrosine based activation motif-containing CEACAM1-related immune receptors in the dog
Abstract
Background: Although the impact of pathogens on the evolution of the mammalian immune system is still under debate, proteins, which both regulate immune responses and serve as cellular receptors for pathogens should be at the forefront of pathogen-driven host evolution. The CEA (carcinoembryonic antigen) gene family codes for such proteins and indeed shows tremendous species-specific variation between human and rodents. Since little is known about the CEA gene family in other lineages of placental mammals, we expected to gain new insights into the evolution of the rapidly diverging CEA family by analyzing the CEA family of the dog.
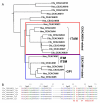
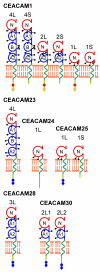
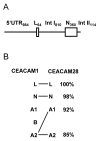
Results: Here we describe the complete CEA gene family in the dog. We found that the gene coding for the ITIM-bearing immunoregulatory molecule CEACAM1 gave rise to a recent expansion of the canine CEA gene family by gene duplication, similar to that previously found in humans and mice. However, while the murine and human CEACAMs (carcinoembryonic antigen-related cell adhesion molecules) are predominantly secreted and GPI-anchored, respectively, in the dog, most of the CEACAMs represent ITAM-bearing transmembrane proteins. One of these proteins, CEACAM28, exhibits nearly complete sequence identity with the ligand-binding N domain of CEACAM1, but antagonizing signaling motifs in the cytoplasmic tail. Comparison of nonsynonymous and synonymous substitutions indicates that the CEACAM28 N domain is under the strongest purifying selection of all canine CEACAM1-related CEACAMs. In addition, CEACAM28 shows a similar expression pattern in resting immune cells and tissues as CEACAM1. However, upon activation CEACAM28 mRNA and CEACAM1 mRNA are differentially regulated.
Conclusion: Thus, CEACAM1 and CEACAM28 are the first paired immune receptors identified within the CEA gene family, which are expressed on T cells and are most likely involved in the fine-tuning of T cell responses. The direction of gene conversion accompanied by purifying selection and expression in immune cells suggests the possibility that CEACAM28 evolved in response to selective pressure imposed by species-specific pathogens.
Figures






References
-
- Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, Hellsten U, Goodstein D, Couronne O, Tran-Gyamfi M, Aerts A, Altherr M, Ashworth L, Bajorek E, Black S, Branscomb E, Caenepeel S, Carrano A, Caoile C, Chan YM, Christensen M, Cleland CA, Copeland A, Dalin E, Dehal P, Denys M, Detter JC, Escobar J, Flowers D, Fotopulos D, Garcia C, Georgescu AM, Glavina T, Gomez M, Gonzales E, Groza M, Hammon N, Hawkins T, Haydu L, Ho I, Huang W, Israni S, Jett J, Kadner K, Kimball H, Kobayashi A, Larionov V, Leem SH, Lopez F, Lou Y, Lowry S, Malfatti S, Martinez D, McCready P, Medina C, Morgan J, Nelson K, Nolan M, Ovcharenko I, Pitluck S, Pollard M, Popkie AP, Predki P, Quan G, Ramirez L, Rash S, Retterer J, Rodriguez A, Rogers S, Salamov A, Salazar A, She X, Smith D, Slezak T, Solovyev V, Thayer N, Tice H, Tsai M, Ustaszewska A, Vo N, Wagner M, Wheeler J, Wu K, Xie G, Yang J, Dubchak I, Furey TS, DeJong P, Dickson M, Gordon D, Eichler EE, Pennacchio LA, Richardson P, Stubbs L, Rokhsar DS, Myers RM, Rubin EM, Lucas SM. The DNA sequence and biology of human chromosome 19. Nature. 2004;428:529–535. doi: 10.1038/nature02399. - DOI - PubMed
-
- Beauchemin N, Draber P, Dveksler G, Gold P, Gray-Owen S, Grunert F, Hammarstrom S, Holmes KV, Karlsson A, Kuroki M, Lin SH, Lucka L, Najjar SM, Neumaier M, Obrink B, Shively JE, Skubitz KM, Stanners CP, Thomas P, Thompson JA, Virji M, von Kleist S, Wagener C, Watt S, Zimmermann W. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

