SH2B1beta (SH2-Bbeta) enhances expression of a subset of nerve growth factor-regulated genes important for neuronal differentiation including genes encoding urokinase plasminogen activator receptor and matrix metalloproteinase 3/10
- PMID: 17947375
- PMCID: PMC2234583
- DOI: 10.1210/me.2007-0384
SH2B1beta (SH2-Bbeta) enhances expression of a subset of nerve growth factor-regulated genes important for neuronal differentiation including genes encoding urokinase plasminogen activator receptor and matrix metalloproteinase 3/10
Abstract
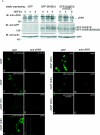
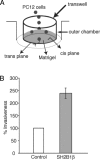
Previous work showed that the adapter protein SH2B adapter protein 1beta (SH2B1) (SH2-B) binds to the activated form of the nerve growth factor (NGF) receptor TrkA and is critical for both NGF-dependent neurite outgrowth and maintenance. To identify SH2B1beta-regulated genes critical for neurite outgrowth, we performed microarray analysis of control PC12 cells and PC12 cells stably overexpressing SH2B1beta (PC12-SH2B1beta) or the dominant-negative SH2B1beta(R555E) [PC12-SH2B1beta(R555E)]. NGF-induced microarray expression of Plaur and Mmp10 genes was greatly enhanced in PC12-SH2B1beta cells, whereas NGF-induced Plaur and Mmp3 expression was substantially depressed in PC12-SH2B1beta(R555E) cells. Plaur, Mmp3, and Mmp10 are among the 12 genes most highly up-regulated after 6 h of NGF. Their protein products [urokinase plasminogen activator receptor (uPAR), matrix metalloproteinase 3 (MMP3), and MMP10] lie in the same pathway of extracellular matrix degradation; uPAR has been shown previously to be critical for NGF-induced neurite outgrowth. Quantitative real-time PCR analysis revealed SH2B1beta enhancement of NGF induction of all three genes and the suppression of NGF induction of all three when endogenous SH2B1 was reduced using short hairpin RNA against SH2B1 and in PC12-SH2B1beta(R555E) cells. NGF-induced levels of uPAR and MMP3/10 and neurite outgrowth through Matrigel (MMP3-dependent) were also increased in PC12-SH2B1beta cells. These results suggest that SH2B1beta stimulates NGF-induced neuronal differentiation at least in part by enhancing expression of a specific subset of NGF-sensitive genes, including Plaur, Mmp3, and/or Mmp10, required for neurite outgrowth.
Figures






References
-
- Thoenen H, Barde YA 1980 Physiology of nerve growth factor. Physiol Rev 60:1284–1335 - PubMed
-
- Levi-Montalcini R 1987 The nerve growth factor 35 years later. Science 237:1154–1162 - PubMed
-
- Oppenheim RW 1991 Cell death during development of the nervous system. Annu Rev Neurosci 14:453–501 - PubMed
-
- Sofroniew MV, Howe CL, Mobley WC 2001 Nerve growth factor signaling, neuroprotection, and neural repair. Annu Rev Neurosci 24:1217–1281 - PubMed
-
- Francis NJ, Landis SC 1999 Cellular and molecular determinants of sympathetic neuron development. Annu Rev Neurosci 22:541–566 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

