Accelerated rate of gene gain and loss in primates
- PMID: 17947411
- PMCID: PMC2147951
- DOI: 10.1534/genetics.107.080077
Accelerated rate of gene gain and loss in primates
Abstract
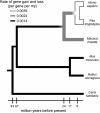
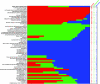
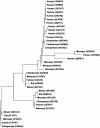
The molecular changes responsible for the evolution of modern humans have primarily been discussed in terms of individual nucleotide substitutions in regulatory or protein coding sequences. However, rates of nucleotide substitution are slowed in primates, and thus humans and chimpanzees are highly similar at the nucleotide level. We find that a third source of molecular evolution, gene gain and loss, is accelerated in primates relative to other mammals. Using a novel method that allows estimation of rate heterogeneity among lineages, we find that the rate of gene turnover in humans is more than 2.5 times faster than in other mammals and may be due to both mutational and selective forces. By reconciling the gene trees for all of the gene families included in the analysis, we are able to independently verify the numbers of inferred duplications. We also use two methods based on the genome assembly of rhesus macaque to further verify our results. Our analyses identify several gene families that have expanded or contracted more rapidly than is expected even after accounting for an overall rate acceleration in primates, including brain-related families that have more than doubled in size in humans. Many of the families showing large expansions also show evidence for positive selection on their nucleotide sequences, suggesting that selection has been important in shaping copy-number differences among mammals. These findings may help explain why humans and chimpanzees show high similarity between orthologous nucleotides yet great morphological and behavioral differences.
Figures
References
-
- Adkins, R. M., A. H. Walton and R. L. Honeycutt, 2003. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol. Phylogenet. Evol. 26: 409–420. - PubMed
-
- Bailey, J. A., Z. P. Gu, R. A. Clark, K. Reinert, R. V. Samonte et al., 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

