Detection of quantitative trait Loci influencing recombination using recombinant inbred lines
- PMID: 17947433
- PMCID: PMC2219516
- DOI: 10.1534/genetics.107.076679
Detection of quantitative trait Loci influencing recombination using recombinant inbred lines
Abstract
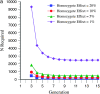
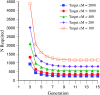
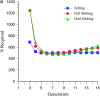
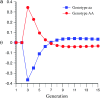
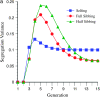
The genetic basis of variation in recombination in higher plants is polygenic and poorly understood, despite its theoretical and practical importance. Here a method of detecting quantitative trait loci (QTL) influencing recombination in recombinant inbred lines (RILs) is proposed that relies upon the fact that genotype data within RILs carry the signature of past recombination. Behavior of the segregational genetic variance in numbers of chromosomal crossovers (recombination) over generations is described for self-, full-sib-, and half-sib-generated RILs with no dominance in true crossovers. This genetic variance, which as a fraction of the total phenotypic variance contributes to the statistical power of the method, was asymptotically greatest with half sibbing, less with sibbing, and least with selfing. The statistical power to detect a recombination QTL declined with diminishing QTL effect, genome target size, and marker density. For reasonably tight marker linkage power was greater with less intense inbreeding for later generations and vice versa for early generations. Generational optima for segregation variance and statistical power were found, whose onset and narrowness varied with marker density and mating design, being more pronounced for looser marker linkage. Application of this method to a maize RIL population derived from inbred lines Mo17 and B73 and developed by selfing suggested two putative QTL (LOD > 2.4) affecting certain chromosomes, and using a canonical transformation another putative QTL was detected. However, permutation tests failed to support their presence (experimentwise alpha = 0.05). Other populations with more statistical power and chosen specifically for recombination QTL segregation would be more effective.
Figures







References
-
- Beavis, W. D., and D. Grant, 1991. A linkage map based on information from four F2 populations of maize (Zea mays L.). Theor. Appl. Genet. 82: 636–644. - PubMed
-
- Brown, H.S., 1961. Differential chiasma frequencies in self-pollinating and cross- pollinating species of the genus Gilia. Aliso 5: 67–81.
-
- Busso, C. F., C. J. Liu, C. T. Hash, J. R. Witcombe, K. M. Devos et al., 1995. Analysis of recombination rate in female and male gametogenesis in pearl millet (Pennisetum glaucum), using RFLP markers. Theor. Appl. Genet. 90: 242–246. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

