Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene
- PMID: 17947439
- PMCID: PMC2219477
- DOI: 10.1534/genetics.107.075598
Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene
Abstract
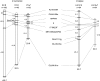
The medaka Oryzias latipes and its two sister species, O. curvinotus and O. luzonensis, possess an XX-XY sex-determination system. The medaka sex-determining gene DMY has been identified on the orthologous Y chromosome [O. latipes linkage group 1 (LG1)] of O. curvinotus. However, DMY has not been discovered in other Oryzias species. These results and molecular phylogeny suggest that DMY was generated recently [approximately 10 million years ago (MYA)] by gene duplication of DMRT1 in a common ancestor of O. latipes and O. curvinotus. We identified seven sex-linked markers from O. luzonensis (sister species of O. curvinotus) and constructed a sex-linkage map. Surprisingly, all seven sex-linked markers were located on an autosomal linkage group (LG12) of O. latipes. As suggested by the phylogenetic tree, the sex chromosomes of O. luzonensis should be "younger" than those of O. latipes. In the lineage leading to O. luzonensis after separation from O. curvinotus approximately 5 MYA, a novel sex-determining gene may have arisen and substituted for DMY. Oryzias species should provide a useful model for evolution of the master sex-determining gene and differentiation of sex chromosomes from autosomes.
Figures




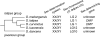
Similar articles
-
Interspecific hybridization between Oryzias latipes and Oryzias curvinotus causes XY sex reversal.J Exp Zool A Comp Exp Biol. 2006 Oct 1;305(10):890-6. doi: 10.1002/jez.a.330. J Exp Zool A Comp Exp Biol. 2006. PMID: 16941651
-
The XX-XY sex-determination system in Oryzias luzonensis and O. mekongensis revealed by the sex ratio of the progeny of sex-reversed fish.Zoolog Sci. 2004 Oct;21(10):1015-8. doi: 10.2108/zsj.21.1015. Zoolog Sci. 2004. PMID: 15514470
-
Identification of the sex-determining locus in the Thai medaka, Oryzias minutillus.Cytogenet Genome Res. 2008;121(2):137-42. doi: 10.1159/000125839. Epub 2008 Jun 9. Cytogenet Genome Res. 2008. PMID: 18544937
-
Sex determination and sex chromosome evolution: insights from medaka.Sex Dev. 2009;3(2-3):88-98. doi: 10.1159/000223074. Epub 2009 Aug 10. Sex Dev. 2009. PMID: 19684454 Review.
-
Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus.Cytogenet Genome Res. 2002;99(1-4):170-7. doi: 10.1159/000071590. Cytogenet Genome Res. 2002. PMID: 12900561 Review.
Cited by
-
Turnover of sex chromosomes and speciation in fishes.Environ Biol Fishes. 2012;94(3):549-558. doi: 10.1007/s10641-011-9853-8. Epub 2011 Jun 4. Environ Biol Fishes. 2012. PMID: 26069393 Free PMC article.
-
Genomic approaches to study genetic and environmental influences on fish sex determination and differentiation.Mar Biotechnol (NY). 2012 Oct;14(5):591-604. doi: 10.1007/s10126-012-9445-4. Epub 2012 Apr 29. Mar Biotechnol (NY). 2012. PMID: 22544374 Free PMC article. Review.
-
Genomic characterization of the Atlantic cod sex-locus.Sci Rep. 2016 Aug 8;6:31235. doi: 10.1038/srep31235. Sci Rep. 2016. PMID: 27499266 Free PMC article.
-
The male and female genomes of golden pompano (Trachinotus ovatus) provide insights into the sex chromosome evolution and rapid growth.J Adv Res. 2024 Nov;65:1-17. doi: 10.1016/j.jare.2023.11.030. Epub 2023 Dec 2. J Adv Res. 2024. PMID: 38043610 Free PMC article.
-
The Y chromosome that lost the male-determining function behaves as an X chromosome in the medaka fish, Oryzias latipes.Genetics. 2008 Aug;179(4):2157-62. doi: 10.1534/genetics.108.090167. Epub 2008 Aug 9. Genetics. 2008. PMID: 18689894 Free PMC article.
References
-
- Charlesworth, D., B. Charlesworth and G. Marais, 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. - PubMed
-
- Devlin, R. H., and Y. Nagahama, 2002. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208: 191–364.
-
- Formacion, M. J., and H. Uwa, 1985. Cytogenetic studies on the origin and species differentiation of the Philippine medaka, Oryzias luzonensis. J. Fish Biol. 27: 285–291.
-
- Foster, J. W., F. E. Brennan, G. K. Hampikian, P. N. Goodfellow, A. H. Sinclair et al., 1992. Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359: 531–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

