Sequence-level population simulations over large genomic regions
- PMID: 17947444
- PMCID: PMC2147962
- DOI: 10.1534/genetics.106.069088
Sequence-level population simulations over large genomic regions
Abstract
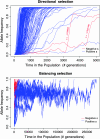
Simulation is an invaluable tool for investigating the effects of various population genetics modeling assumptions on resulting patterns of genetic diversity, and for assessing the performance of statistical techniques, for example those designed to detect and measure the genomic effects of selection. It is also used to investigate the effectiveness of various design options for genetic association studies. Backward-in-time simulation methods are computationally efficient and have become widely used since their introduction in the 1980s. The forward-in-time approach has substantial advantages in terms of accuracy and modeling flexibility, but at greater computational cost. We have developed flexible and efficient simulation software and a rescaling technique to aid computational efficiency that together allow the simulation of sequence-level data over large genomic regions in entire diploid populations under various scenarios for demography, mutation, selection, and recombination, the latter including hotspots and gene conversion. Our forward evolution of genomic regions (FREGENE) software is freely available from www.ebi.ac.uk/projects/BARGEN together with an ancillary program to generate phenotype labels, either binary or quantitative. In this article we discuss limitations of coalescent-based simulation, introduce the rescaling technique that makes large-scale forward-in-time simulation feasible, and demonstrate the utility of various features of FREGENE, many not previously available.
Figures


References
-
- Crawford, D. C., T. Bhangale, N. Li, G. Hellenthal, M. J. Rieder et al., 2004. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36: 700–706. - PubMed
-
- Ewens, W., 2004. Mathematical Population Genetics: I. Theoretical Introduction. Springer-Verlag, New York.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

