The structure of FtsZ filaments in vivo suggests a force-generating role in cell division
- PMID: 17948052
- PMCID: PMC2080809
- DOI: 10.1038/sj.emboj.7601895
The structure of FtsZ filaments in vivo suggests a force-generating role in cell division
Abstract
In prokaryotes, FtsZ (the filamentous temperature sensitive protein Z) is a nearly ubiquitous GTPase that localizes in a ring at the leading edge of constricting plasma membranes during cell division. Here we report electron cryotomographic reconstructions of dividing Caulobacter crescentus cells wherein individual arc-like filaments were resolved just underneath the inner membrane at constriction sites. The filaments' position, orientation, time of appearance, and resistance to A22 all suggested that they were FtsZ. Predictable changes in the number, length, and distribution of filaments in cells where the expression levels and stability of FtsZ were altered supported that conclusion. In contrast to the thick, closed-ring-like structure suggested by fluorescence light microscopy, throughout the constriction process the Z-ring was seen here to consist of just a few short (approximately 100 nm) filaments spaced erratically near the division site. Additional densities connecting filaments to the cell wall, occasional straight segments, and abrupt kinks were also seen. An 'iterative pinching' model is proposed wherein FtsZ itself generates the force that constricts the membrane in a GTP-hydrolysis-driven cycle of polymerization, membrane attachment, conformational change, depolymerization, and nucleotide exchange.
Figures




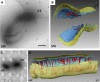




Similar articles
-
Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division.Elife. 2014 Dec 9;3:e04601. doi: 10.7554/eLife.04601. Elife. 2014. PMID: 25490152 Free PMC article.
-
Short FtsZ filaments can drive asymmetric cell envelope constriction at the onset of bacterial cytokinesis.EMBO J. 2017 Jun 1;36(11):1577-1589. doi: 10.15252/embj.201696235. Epub 2017 Apr 24. EMBO J. 2017. PMID: 28438890 Free PMC article.
-
The intrinsically disordered C-terminal linker of FtsZ regulates protofilament dynamics and superstructure in vitro.J Biol Chem. 2017 Dec 15;292(50):20509-20527. doi: 10.1074/jbc.M117.809939. Epub 2017 Oct 31. J Biol Chem. 2017. PMID: 29089389 Free PMC article.
-
FtsZ-ring Architecture and Its Control by MinCD.Subcell Biochem. 2017;84:213-244. doi: 10.1007/978-3-319-53047-5_7. Subcell Biochem. 2017. PMID: 28500527 Review.
-
Form and function of the bacterial cytokinetic ring.Curr Opin Cell Biol. 2014 Feb;26:19-27. doi: 10.1016/j.ceb.2013.08.006. Epub 2013 Sep 25. Curr Opin Cell Biol. 2014. PMID: 24529242 Review.
Cited by
-
Multidimensional view of the bacterial cytoskeleton.J Bacteriol. 2013 Apr;195(8):1627-36. doi: 10.1128/JB.02194-12. Epub 2013 Feb 15. J Bacteriol. 2013. PMID: 23417493 Free PMC article. Review.
-
Probing for Binding Regions of the FtsZ Protein Surface through Site-Directed Insertions: Discovery of Fully Functional FtsZ-Fluorescent Proteins.J Bacteriol. 2016 Dec 13;199(1):e00553-16. doi: 10.1128/JB.00553-16. Print 2017 Jan 1. J Bacteriol. 2016. PMID: 27795325 Free PMC article.
-
Geometric constrains for detecting short actin filaments by cryogenic electron tomography.PMC Biophys. 2010 Mar 5;3(1):6. doi: 10.1186/1757-5036-3-6. PMC Biophys. 2010. PMID: 20214767 Free PMC article.
-
Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli.J Bacteriol. 2011 Mar;193(6):1405-13. doi: 10.1128/JB.01258-10. Epub 2011 Jan 7. J Bacteriol. 2011. PMID: 21216995 Free PMC article.
-
FtsZ polymerization assays: simple protocols and considerations.J Vis Exp. 2013 Nov 16;(81):e50844. doi: 10.3791/50844. J Vis Exp. 2013. PMID: 24300445 Free PMC article.
References
-
- Addinall SG, Lutkenhaus J (1996) FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol 22: 231–237 - PubMed
-
- Ben-Yehuda S, Losick R (2002) Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109: 257–266 - PubMed
-
- Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

