Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring
- PMID: 17949071
- PMCID: PMC2564819
- DOI: 10.1021/ci700271z
Search for non-nucleoside inhibitors of HIV-1 reverse transcriptase using chemical similarity, molecular docking, and MM-GB/SA scoring
Abstract
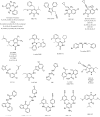
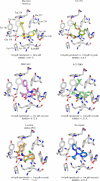
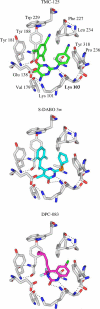
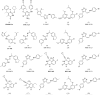
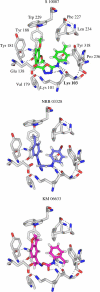
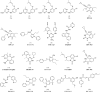
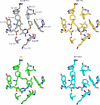
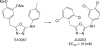
A virtual screening protocol has been applied to seek non-nucleoside inhibitors of HIV-1 reverse transcriptase (NNRTIs) and its K103N mutant. First, a chemical similarity search on the Maybridge library was performed using known NNRTIs as reference structures. The top-ranked molecules obtained from this procedure plus 26 known NNRTIs were then docked into the binding sites of the wild-type reverse transcriptase (HIV-RT) and its K103N variant (K103N-RT) using Glide 3.5. The top-ranked 100 compounds from the docking for both proteins were post-scored with a procedure using molecular mechanics and continuum solvation (MM-GB/SA). The validity of the virtual screening protocol was supported by (i) testing of the MM-GB/SA procedure, (ii) agreement between predicted and crystallographic binding poses, (iii) recovery of known potent NNRTIs at the top of both rankings, and (iv) identification of top-scoring library compounds that are close in structure to recently reported NNRTI HTS hits. However, purchase and assaying of selected top-scoring compounds from the library failed to yield active anti-HIV agents. Nevertheless, the highest-ranked database compound, S10087, was pursued as containing a potentially viable core. Subsequent synthesis and assaying of S10087 analogues proposed by further computational analysis yielded anti-HIV agents with EC50 values as low as 310 nM. Thus, with the aid of computational tools, it was possible to evolve a false positive into a true active.
Figures



References
-
- 2006 AIDS Epidemic Update. UNAIDS; Geneva: 2006.
-
- De Clercq E. New anti-HIV agents and targets. Med. Res. Rev. 2002;22:531–565. - PubMed
-
- Katz RA, Skalka AM. The Retroviral Enzymes. Annu. Rev. Biochem. 1994;63:133–173. - PubMed
-
- Tantillo C, Ding J, Jacobo-Molina A, Nanni RG, Boyer PL, Hughes SH, Pauwels R, Andries K, Janssen PAJ, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase: implications for mechanisms of drug inhibition and resistance. J Mol Biol. 1994;243:369–387. - PubMed
-
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

