Effects of iron loading on muscle: genome-wide mRNA expression profiling in the mouse
- PMID: 17949489
- PMCID: PMC2151772
- DOI: 10.1186/1471-2164-8-379
Effects of iron loading on muscle: genome-wide mRNA expression profiling in the mouse
Abstract
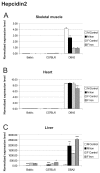
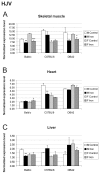
Background: Hereditary hemochromatosis (HH) encompasses genetic disorders of iron overload characterized by deficient expression or function of the iron-regulatory hormone hepcidin. Mutations in 5 genes have been linked to this disease: HFE, TFR2 (encoding transferrin receptor 2), HAMP (encoding hepcidin), SLC40A1 (encoding ferroportin) and HJV (encoding hemojuvelin). Hepcidin inhibits iron export from cells into plasma. Hemojuvelin, an upstream regulator of hepcidin expression, is expressed in mice mainly in the heart and skeletal muscle. It has been suggested that soluble hemojuvelin shed by the muscle might reach the liver to influence hepcidin expression. Heart muscle is one of the target tissues affected by iron overload, with resultant cardiomyopathy in some HH patients. Therefore, we investigated the effect of iron overload on gene expression in skeletal muscle and heart using Illuminatrade mark arrays containing over 47,000 probes. The most apparent changes in gene expression were confirmed using real-time RT-PCR.
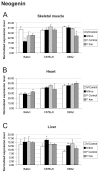
Results: Genes with up-regulated expression after iron overload in both skeletal and heart muscle included angiopoietin-like 4, pyruvate dehydrogenase kinase 4 and calgranulin A and B. The expression of transferrin receptor, heat shock protein 1B and DnaJ homolog B1 were down-regulated by iron in both muscle types. Two potential hepcidin regulatory genes, hemojuvelin and neogenin, showed no clear change in expression after iron overload.
Conclusion: Microarray analysis revealed iron-induced changes in the expression of several genes involved in the regulation of glucose and lipid metabolism, transcription and cellular stress responses. These may represent novel connections between iron overload and pathological manifestations of HH such as cardiomyopathy and diabetes.
Figures



Similar articles
-
Hepatic iron metabolism gene expression profiles in HFE associated hereditary hemochromatosis.Blood Cells Mol Dis. 2007 Jan-Feb;38(1):37-44. doi: 10.1016/j.bcmd.2006.09.008. Epub 2006 Nov 13. Blood Cells Mol Dis. 2007. PMID: 17098454
-
Hereditary hemochromatosis: mutations in genes involved in iron homeostasis in Brazilian patients.Blood Cells Mol Dis. 2011 Apr 15;46(4):302-7. doi: 10.1016/j.bcmd.2011.02.008. Blood Cells Mol Dis. 2011. PMID: 21411349
-
Inherited Disorders of Iron Overload.Front Nutr. 2018 Oct 29;5:103. doi: 10.3389/fnut.2018.00103. eCollection 2018. Front Nutr. 2018. PMID: 30420953 Free PMC article. Review.
-
Hepatic expression of hemochromatosis genes in two mouse strains after phlebotomy and iron overload.Haematologica. 2005 Sep;90(9):1161-7. Haematologica. 2005. PMID: 16154838
-
The mechanisms of systemic iron homeostasis and etiology, diagnosis, and treatment of hereditary hemochromatosis.Int J Hematol. 2018 Jan;107(1):31-43. doi: 10.1007/s12185-017-2365-3. Epub 2017 Nov 13. Int J Hematol. 2018. PMID: 29134618 Review.
Cited by
-
An ex vivo model for studying hepatic schistosomiasis and the effect of released protein from dying eggs.PLoS Negl Trop Dis. 2015 May 12;9(5):e0003760. doi: 10.1371/journal.pntd.0003760. eCollection 2015 May. PLoS Negl Trop Dis. 2015. PMID: 25965781 Free PMC article.
-
Evaluation of Different Normalization and Analysis Procedures for Illumina Gene Expression Microarray Data Involving Small Changes.Microarrays (Basel). 2013 May 21;2(2):131-52. doi: 10.3390/microarrays2020131. Microarrays (Basel). 2013. PMID: 27605185 Free PMC article.
-
Global transcriptional response to Hfe deficiency and dietary iron overload in mouse liver and duodenum.PLoS One. 2009 Sep 29;4(9):e7212. doi: 10.1371/journal.pone.0007212. PLoS One. 2009. PMID: 19787063 Free PMC article.
-
Orchestrated regulation of iron trafficking proteins in the kidney during iron overload facilitates systemic iron retention.PLoS One. 2018 Oct 15;13(10):e0204471. doi: 10.1371/journal.pone.0204471. eCollection 2018. PLoS One. 2018. PMID: 30321179 Free PMC article.
-
Association between aluminum and iron exposure in maternal blood and umbilical cord blood and congenital heart defects in children.PeerJ. 2024 Jan 22;12:e16755. doi: 10.7717/peerj.16755. eCollection 2024. PeerJ. 2024. PMID: 38274332 Free PMC article.
References
-
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R, Jr., Ellis MC, Fullan A, Hinton LM, Jones NL, Kimmel BE, Kronmal GS, Lauer P, Lee VK, Loeb DB, Mapa FA, McClelland E, Meyer NC, Mintier GA, Moeller N, Moore T, Morikang E, Prass CE, Quintana L, Starnes SM, Schatzman RC, Brunke KJ, Drayna DT, Risch NJ, Bacon BR, Wolff RK. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. - DOI - PubMed
-
- Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

