Proteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells
- PMID: 17949499
- PMCID: PMC2148065
- DOI: 10.1186/1471-2164-8-380
Proteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells
Abstract
Background: Recently, we demonstrated that human mesenchymal stem cells (hMSC) stimulated with dexamethazone undergo gene focusing during osteogenic differentiation (Stem Cells Dev 14(6): 1608-20, 2005). Here, we examine the protein expression profiles of three additional populations of hMSC stimulated to undergo osteogenic differentiation via either contact with pro-osteogenic extracellular matrix (ECM) proteins (collagen I, vitronectin, or laminin-5) or osteogenic media supplements (OS media). Specifically, we annotate these four protein expression profiles, as well as profiles from naïve hMSC and differentiated human osteoblasts (hOST), with known gene ontologies and analyze them as a tensor with modes for the expressed proteins, gene ontologies, and stimulants.
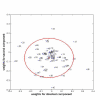
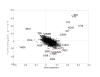
Results: Direct component analysis in the gene ontology space identifies three components that account for 90% of the variance between hMSC, osteoblasts, and the four stimulated hMSC populations. The directed component maps the differentiation stages of the stimulated stem cell populations along the differentiation axis created by the difference in the expression profiles of hMSC and hOST. Surprisingly, hMSC treated with ECM proteins lie closer to osteoblasts than do hMSC treated with OS media. Additionally, the second component demonstrates that proteomic profiles of collagen I- and vitronectin-stimulated hMSC are distinct from those of OS-stimulated cells. A three-mode tensor analysis reveals additional focus proteins critical for characterizing the phenotypic variations between naïve hMSC, partially differentiated hMSC, and hOST.
Conclusion: The differences between the proteomic profiles of OS-stimulated hMSC and ECM-hMSC characterize different transitional phenotypes en route to becoming osteoblasts. This conclusion is arrived at via a three-mode tensor analysis validated using hMSC plated on laminin-5.
Figures



References
-
- Le Blanc K, Pittenger M. Mesenchymal stem cells: progress toward promise. Cytotherapy. 2005;7:36–45. - PubMed
-
- Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568–584. - PubMed
-
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. - PubMed
-
- Aubin JE. Advances in the osteoblast lineage. Biochem Cell Biol. 1998;76:899–910. - PubMed
-
- Smith E, Redman RA, Logg CR, Coetzee GA, Kasahara N, Frenkel B. Glucocorticoids inhibit developmental stage-specific osteoblast cell cycle. Dissociation of cyclin A-cyclin-dependent kinase 2 from E2F4-p130 complexes. J Biol Chem. 2000;275:19992–20001. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

