GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans
- PMID: 17950010
- PMCID: PMC2683589
- DOI: 10.1016/j.fgb.2007.08.006
GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans
Abstract
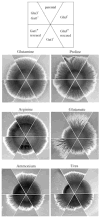
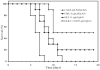
The function of GLN3, a GATA factor encoding gene, in nitrogen metabolism of Candida albicans was examined. GLN3 null mutants had reduced growth rates on multiple nitrogen sources. More severe growth defects were observed in mutants lacking both GLN3 and GAT1, a second GATA factor gene. GLN3 was an activator of two genes involved in ammonium assimilation, GDH3, encoding NADP-dependent glutamate dehydrogenase, and MEP2, which encodes an ammonium permease. GAT1 contributed to MEP2 expression, but not that of GDH3. A putative general amino acid permease gene, GAP2, was also activated by both GLN3 and GAT1, but activation by GLN3 was nitrogen source dependent. GLN3 was constitutively expressed, but GAT1 expression varied with nitrogen source and was reduced 2- to 3-fold in gln3 mutants. Both gln3 and gat1 mutants exhibited reduced sensitivity to rapamycin, suggesting they function downstream of TOR kinase. Hyphae formation by gln3 and gat1 mutants differed in relation to nitrogen source. The gln3 mutants formed hyphae on several nitrogen sources, but not ammonium or urea, suggesting a defect in ammonium assimilation. Virulence of gln3 mutants was reduced in a murine model of disseminated disease. We conclude that GLN3 has a broad role in nitrogen metabolism, partially overlapping, but distinct from that of GAT1, and that its function is important for the ability of C. albicans to survive within the host environment.
Figures




References
-
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient- regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Bertram PG, et al. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–33. - PubMed
-
- Biswas K, Morschhauser J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol. 2005;56:649–69. - PubMed
-
- Boeke JD, et al. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

