Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes
- PMID: 17950269
- PMCID: PMC2170521
- DOI: 10.1016/j.ydbio.2007.09.018
Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes
Abstract
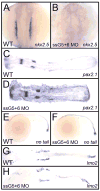
An outstanding problem in vertebrate development has been to define the genetic program that specifies the cardiomyocyte lineage. It has been a challenge to define the transcription factors that control specification, since candidate gene knockouts typically cause rather complex morphogenetic defects. In contrast, Drosophila genetics identified single transcription factors that are essential for specification of cardiomyocytes from uncommitted mesoderm. For those vertebrate orthologs, it has been considered that paralogous family members might compensate for the loss-of-function of individual genes. However, this hypothesis had not been formally tested. In zebrafish, defects in gata5 can lead to a loss of myocardial tissue, but most embryos depleted for any single vertebrate Gata4/5/6 transcription factor develop a cardiac morphogenetic defect, and cardiomyocytes are specified and differentiate. Here we show that in zebrafish the gata5 and gata6 genes are redundant for specification of cardiomyocytes. Embryos depleted of these two gene products are heartless. Restoring either gene product is sufficient to rescue cardiomyocyte specification. In contrast, embryos depleted of Gata4 and Gata6, or Gata4 and Gata5, develop defective heart tubes. Our study identifies a specific pair of vertebrate transcription factor paralogs that is essential for cardiomyocyte specification.
Figures




Similar articles
-
Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling.Dev Dyn. 2011 Mar;240(3):649-62. doi: 10.1002/dvdy.22570. Epub 2011 Feb 8. Dev Dyn. 2011. PMID: 21305652
-
GATA5 interacts with GATA4 and GATA6 in outflow tract development.Dev Biol. 2011 Oct 15;358(2):368-78. doi: 10.1016/j.ydbio.2011.07.037. Epub 2011 Aug 4. Dev Biol. 2011. PMID: 21839733
-
Tmem88a mediates GATA-dependent specification of cardiomyocyte progenitors by restricting WNT signaling.Development. 2013 Sep;140(18):3787-98. doi: 10.1242/dev.093567. Epub 2013 Jul 31. Development. 2013. PMID: 23903195 Free PMC article.
-
Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way?Int J Mol Sci. 2022 May 9;23(9):5255. doi: 10.3390/ijms23095255. Int J Mol Sci. 2022. PMID: 35563646 Free PMC article. Review.
-
Using the zebrafish model to study GATA transcription factors.Semin Cell Dev Biol. 2005 Feb;16(1):95-106. doi: 10.1016/j.semcdb.2004.10.004. Epub 2004 Dec 10. Semin Cell Dev Biol. 2005. PMID: 15659344 Review.
Cited by
-
The centrosomal protein 83 (CEP83) regulates human pluripotent stem cell differentiation toward the kidney lineage.Elife. 2022 Oct 12;11:e80165. doi: 10.7554/eLife.80165. Elife. 2022. PMID: 36222666 Free PMC article.
-
Modeling Human Cardiac Arrhythmias: Insights from Zebrafish.J Cardiovasc Dev Dis. 2022 Jan 5;9(1):13. doi: 10.3390/jcdd9010013. J Cardiovasc Dev Dis. 2022. PMID: 35050223 Free PMC article. Review.
-
Common genetic control of haemangioblast and cardiac development in zebrafish.Development. 2009 May;136(9):1465-74. doi: 10.1242/dev.032748. Epub 2009 Mar 18. Development. 2009. PMID: 19297410 Free PMC article.
-
Genome-wide transcriptomics analysis identifies sox7 and sox18 as specifically regulated by gata4 in cardiomyogenesis.Dev Biol. 2018 Feb 1;434(1):108-120. doi: 10.1016/j.ydbio.2017.11.017. Epub 2017 Dec 8. Dev Biol. 2018. PMID: 29229250 Free PMC article.
-
Specificity, redundancy and dosage thresholds among gata4/5/6 genes during zebrafish cardiogenesis.Biol Open. 2020 Jun 24;9(6):bio053611. doi: 10.1242/bio.053611. Biol Open. 2020. PMID: 32580940 Free PMC article.
References
-
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. - PubMed
-
- Frasch M. Intersecting signalling and transcriptional pathways in Drosophila heart specification. Semin Cell Dev Biol. 1999;10:61–71. - PubMed
-
- Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–49. - PubMed
-
- Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124:2387–95. - PubMed
-
- Heicklen-Klein A, Evans T. T-box binding sites are required for activity of a cardiac GATA-4 enhancer. Dev Biol. 2004;267:490–504. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

