Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands
- PMID: 17951380
- PMCID: PMC2223734
- DOI: 10.1128/JB.01000-07
Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands
Abstract
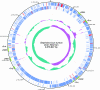
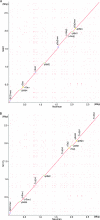
Strains of Staphylococcus aureus, an important human pathogen, display up to 20% variability in their genome sequence, and most sequence information is available for human clinical isolates that have not been subjected to genetic analysis of virulence attributes. S. aureus strain Newman, which was also isolated from a human infection, displays robust virulence properties in animal models of disease and has already been extensively analyzed for its molecular traits of staphylococcal pathogenesis. We report here the complete genome sequence of S. aureus Newman, which carries four integrated prophages, as well as two large pathogenicity islands. In agreement with the view that S. aureus Newman prophages contribute important properties to pathogenesis, fewer virulence factors are found outside of the prophages than for the highly virulent strain MW2. The absence of drug resistance genes reflects the general antibiotic-susceptible phenotype of S. aureus Newman. Phylogenetic analyses reveal clonal relationships between the staphylococcal strains Newman, COL, NCTC8325, and USA300 and a greater evolutionary distance to strains MRSA252, MW2, MSSA476, N315, Mu50, JH1, JH9, and RF122. However, polymorphism analysis of two large pathogenicity islands distributed among these strains shows that the two islands were acquired independently from the evolutionary pathway of the chromosomal backbones of staphylococcal genomes. Prophages and pathogenicity islands play central roles in S. aureus virulence and evolution.
Figures


References
-
- Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. - PubMed
-
- Baba, T., F. Takeuchi, M. Kuroda, T. Ito, H. Yuzawa, and K. Hiramatsu. 2003. The genome of Staphylococcus aureus, p. 66-153. In D. Al'Aladeen and K. Hiramatsu (ed.), The Staphylococcus aureus: molecular and clinical aspects. Ellis Harwood, London, United Kingdom.
-
- Bae, T., T. Baba, K. Hiramatsu, and O. Schneewind. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 621035-1047. - PubMed
-
- Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of acquired methicillin-resistant Staphylococcus aureus. Lancet 367731-739. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

