Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling
- PMID: 17951583
- PMCID: PMC2710213
- DOI: 10.1093/aob/mcm252
Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: with deletion mutants and expression profiling
Abstract
Background and aims: Understanding Ph1, a dominant homoeologous chromosome pairing suppressor locus on the long arm of chromosome 5B in wheat Triticum aestivum L., is the core of the investigation in this article. The Ph1 locus restricts chromosome pairing and recombination at meiosis to true homologues. The importance of wheat as a crop and the need to exploit its wild relatives as donors for economically important traits in wheat breeding programmes is the main drive to uncover the mechanism of the Ph1 locus and regulate its activity.
Methods: Following the molecular genetic characterization of the Ph1 locus, five additional deletion mutants covering the region have been identified. In addition, more bacterial artificial chromosomes (BACs) were sequenced and analysed to elucidate the complexity of this locus. A semi-quantitative RT-PCR was used to compare the expression profiles of different genes in the 5B region containing the Ph1 locus with their homoeologues on 5A and 5D. PCR products were cloned and sequenced to identify the gene from which they were derived.
Key results: Deletion mutants and expression profiling of genes in the region containing the Ph1 locus on 5B has further restricted Ph1 to a cluster of cdk-like genes. Bioinformatic analysis of the cdk-like genes revealed their close homology to the checkpoint kinase Cdk2 from humans. Cdk2 is involved in the initiation of replication and is required in early meiosis. Expression profiling has revealed that the cdk-like gene cluster is unique within the region analysed on 5B in that these genes are transcribed. Deletion of the cdk-like locus on 5B results in activation of transcription of functional cdk-like copies on 5A and 5D. Thus the cdk locus on 5B is dominant to those on 5A and 5D in determining the overall activity, which will be dependent on a complex interplay between transcription from non-functional and functional cdk-like genes.
Conclusions: The Ph1 locus has been defined to a cdk-like gene cluster related to Cdk2 in humans, a master checkpoint gene involved in the initiation of replication and required for early meiosis.
Figures







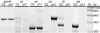
References
-
- Allouis S, Moore G, Bellec A, Sharp R, Faivre Rampant P, Mortimer K, et al. Construction and characterisation of a hexaploid wheat (Triticum aestivum L.) BAC library from the reference germplasm ‘Chinese Spring. Cereal Research Communications. 2003;31:331–338.
-
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. Centromere behaviour in wheat with high and low homoeologous chromosomal pairing. Chromosoma. 1997;106:327–333. - PubMed
-
- Ashley T, Walpita D, de Rooij DG. Localisation of two mammalian cyclin dependent kinases during mammalian meiosis. Journal of Cell Science. 2001;114:685–693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

