Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays
- PMID: 17952125
- PMCID: PMC2492386
- DOI: 10.1038/sj.onc.1210832
Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays
Abstract
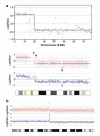
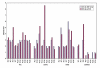
To identify genomic abnormalities characteristic of pancreatic ductal adenocarcinoma (PDAC) in vivo, a panel of 27 microdissected PDAC specimens were analysed using high-density microarrays representing approximately 116 000 single nucleotide polymorphism (SNP) loci. We detected frequent gains of 1q, 2, 3, 5, 7p, 8q, 11, 14q and 17q (> or =78% of cases), and losses of 1p, 3p, 6, 9p, 13q, 14q, 17p and 18q (> or =44%). Although the results were comparable with those from array CGH, regions of those genetic changes were defined more accurately by SNP arrays. Integrating the Ensembl public data, we have generated 'gene' copy number indices that facilitate the search for novel candidates involved in pancreatic carcinogenesis. Copy numbers in a subset of the genes were validated using quantitative real-time PCR. The SKAP2/SCAP2 gene (7p15.2), which belongs to the src family kinases, was most frequently (63%) amplified in our sample set and its recurrent overexpression (67%) was confirmed by reverse transcription-PCR. Furthermore, fluorescence in situ hybridization and in situ RNA hybridization analyses for this gene have demonstrated a significant correlation between DNA copy number and mRNA expression level in an independent sample set (P<0.001). These findings indicate that the dysregulation of SKAP2/SCAP2, which is mostly caused by its increased gene copy number, is likely to be associated with the development of PDAC.
Figures


References
-
- Andersen CL, Wiuf C, Kruhoffer M, Korsgaard M, Laurberg S, Orntoft TF. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48. - PubMed
-
- Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. - PubMed
-
- Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, Schmiegel W, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. - PubMed
-
- Calhoun ES, Hucl T, Gallmeier E, West KM, Arking DE, Maitra A, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–7928. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

