Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies
- PMID: 17953491
- PMCID: PMC2039766
- DOI: 10.1371/journal.pgen.0030190
Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies
Abstract
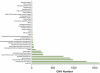
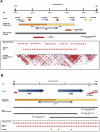
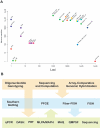
Genome-wide association scans (GWASs) using single nucleotide polymorphisms (SNPs) have been completed successfully for several common disorders and have detected over 30 new associations. Considering the large sample sizes and genome-wide SNP coverage of the scans, one might have expected many of the common variants underpinning the genetic component of various disorders to have been identified by now. However, these studies have not evaluated the contribution of other forms of genetic variation, such as structural variation, mainly in the form of copy number variants (CNVs). Known CNVs account for over 15% of the assembled human genome sequence. Since CNVs are not easily tagged by SNPs, might have a wide range of copy number variability, and often fall in genomic regions not well covered by whole-genome arrays or not genotyped by the HapMap project, current GWASs have largely missed the contribution of CNVs to complex disorders. In fact, some CNVs have already been reported to show association with several complex disorders using candidate gene/region approaches, underpinning the importance of regions not investigated in current GWASs. This reveals the need for new generation arrays (some already in the market) and the use of tailored approaches to explore the full dimension of genome variability beyond the single nucleotide scale.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



References
-
- Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, et al. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. - PubMed
-
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. - PubMed
-
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

