AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape
- PMID: 17954573
- PMCID: PMC2118485
- DOI: 10.1084/jem.20071261
AIDS virus specific CD8+ T lymphocytes against an immunodominant cryptic epitope select for viral escape
Abstract
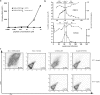
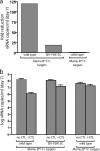
Cryptic major histocompatibility complex class I epitopes have been detected in several pathogens, but their importance in the immune response to AIDS viruses remains unknown. Here, we show that Mamu-B*17(+) simian immunodeficiency virus (SIV)mac239-infected rhesus macaques that spontaneously controlled viral replication consistently made strong CD8(+) T lymphocyte (CD8-TL) responses against a cryptic epitope, RHLAFKCLW (cRW9). Importantly, cRW9-specific CD8-TL selected for viral variation in vivo and effectively suppressed SIV replication in vitro, suggesting that they might play a key role in the SIV-specific response. The discovery of an immunodominant CD8-TL response in elite controller macaques against a cryptic epitope suggests that the AIDS virus-specific cellular immune response is likely far more complex than is generally assumed.
Figures



References
-
- Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

