Differences between homologous alleles of olfactory receptor genes require the Polycomb Group protein Eed
- PMID: 17954609
- PMCID: PMC2064763
- DOI: 10.1083/jcb.200706053
Differences between homologous alleles of olfactory receptor genes require the Polycomb Group protein Eed
Abstract
A number of mammalian genes are expressed from only one of the two homologous chromosomes, selected at random in each cell. These include genes subject to X-inactivation, olfactory receptor (OR) genes, and several classes of immune system genes. The means by which monoallelic expression is established are only beginning to be understood. Using a cytological assay, we show that the two homologous alleles of autosomal random monoallelic loci differ from each other in embryonic stem (ES) cells, before establishment of monoallelic expression. The Polycomb Group gene Eed is required to establish this distinctive behavior. In addition, we found that when Eed mutant ES cells are differentiated, they fail to establish asynchronous replication timing at OR loci. These results suggest a common mechanism for random monoallelic expression on autosomes and the X chromosome, and implicate Eed in establishing differences between homologous OR loci before and after differentiation.
Figures
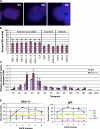
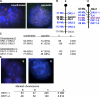
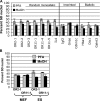

References
-
- Chess, A., I. Simon, H. Cedar, and R. Axel. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. - PubMed
-
- DeChiara, T.M., E.J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 64:849–859. - PubMed
-
- Ensminger, A.W., and A. Chess. 2004. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum. Mol. Genet. 13:651–658. - PubMed

