The transcriptional cycle of HIV-1 in real-time and live cells
- PMID: 17954611
- PMCID: PMC2064765
- DOI: 10.1083/jcb.200706018
The transcriptional cycle of HIV-1 in real-time and live cells
Abstract
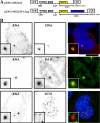
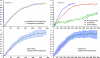
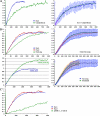
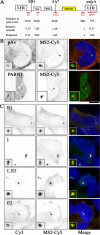
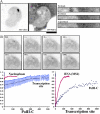
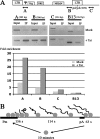
RNA polymerase II (RNAPII) is a fundamental enzyme, but few studies have analyzed its activity in living cells. Using human immunodeficiency virus (HIV) type 1 reporters, we study real-time messenger RNA (mRNA) biogenesis by photobleaching nascent RNAs and RNAPII at specific transcription sites. Through modeling, the use of mutant polymerases, drugs, and quantitative in situ hybridization, we investigate the kinetics of the HIV-1 transcription cycle. Initiation appears efficient because most polymerases demonstrate stable gene association. We calculate an elongation rate of approximately 1.9 kb/min, and, surprisingly, polymerases remain at transcription sites 2.5 min longer than nascent RNAs. With a total polymerase residency time estimated at 333 s, 114 are assigned to elongation, and 63 are assigned to 3'-end processing and/or transcript release. However, mRNAs were released seconds after polyadenylation onset, and analysis of polymerase density by chromatin immunoprecipitation suggests that they pause or lose processivity after passing the polyA site. The strengths and limitations of this kinetic approach to analyze mRNA biogenesis in living cells are discussed.
Figures

References
-
- Ashe, M., P. Griffin, W. James, and N. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008–3025. - PubMed
-
- Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 13:22–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

