Molecular imaging of membrane interfaces reveals mode of beta-glucosidase activation by saposin C
- PMID: 17954913
- PMCID: PMC2077267
- DOI: 10.1073/pnas.0704998104
Molecular imaging of membrane interfaces reveals mode of beta-glucosidase activation by saposin C
Abstract
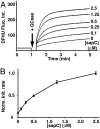
Acid beta-glucosidase (GCase) is a soluble lysosomal enzyme responsible for the hydrolysis of glucose from glucosylceramide and requires activation by the small nonenzymatic protein saposin C (sapC) to gain access to the membrane-embedded glycosphingolipid substrate. We have used in situ atomic force microscopy (AFM) with simultaneous confocal and epifluorescence microscopies to investigate the interactions of GCase and sapC with lipid bilayers. GCase binds to sites on membranes transformed by sapC, and enzyme activity occurs at loci containing both GCase and sapC. Using FRET, we establish the presence of GCase/sapC and GCase/product contacts in the bilayer. These data support a mechanism in which sapC locally alters regions of bilayer for subsequent attack by the enzyme in stably bound protein complexes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Futerman AH, van Meer G. Nat Rev Mol Cell Biol. 2004;5:554–565. - PubMed
-
- Kolter T, Sandhoff K. Annu Rev Cell Dev Biol. 2005;21:81–103. - PubMed
-
- Grabowski GA, Barton NW, Pastores G, Dambrosia JM, Banerjee TK, McKee MA, Parker C, Schiffmann R, Hill SC, Brady RO. Ann Intern Med. 1995;122:33–39. - PubMed
-
- Sandhoff K, Kolter T. Trends Cell Biol. 1996;6:98–103. - PubMed
-
- Vaccaro AM, Salvioli R, Tatti M, Ciaffoni F. Neurochem Res. 1999;24:307–314. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

