Recycling of eukaryotic posttermination ribosomal complexes
- PMID: 17956730
- PMCID: PMC2651563
- DOI: 10.1016/j.cell.2007.08.041
Recycling of eukaryotic posttermination ribosomal complexes
Abstract
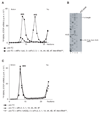
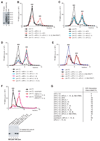
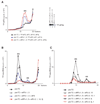
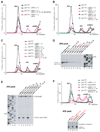
After translational termination, mRNA and P site deacylated tRNA remain associated with ribosomes in posttermination complexes (post-TCs), which must therefore be recycled by releasing mRNA and deacylated tRNA and by dissociating ribosomes into subunits. Recycling of bacterial post-TCs requires elongation factor EF-G and a ribosome recycling factor RRF. Eukaryotes do not encode a RRF homolog, and their mechanism of ribosomal recycling is unknown. We investigated eukaryotic recycling using post-TCs assembled on a model mRNA encoding a tetrapeptide followed by a UAA stop codon and report that initiation factors eIF3, eIF1, eIF1A, and eIF3j, a loosely associated subunit of eIF3, can promote recycling of eukaryotic post-TCs. eIF3 is the principal factor that promotes splitting of posttermination ribosomes into 60S subunits and tRNA- and mRNA-bound 40S subunits. Its activity is enhanced by eIFs 3j, 1, and 1A. eIF1 also mediates release of P site tRNA, whereas eIF3j ensures subsequent dissociation of mRNA.
Figures


References
-
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. - PubMed
-
- Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. - PubMed
-
- Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. - PubMed
-
- Fraser CS, Berry KE, Hershey JWB, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell. 2007;26:811–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

