Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE
- PMID: 17956990
- PMCID: PMC2077256
- DOI: 10.1073/pnas.0705417104
Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE
Abstract
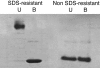
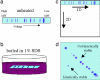
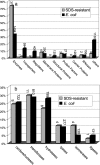
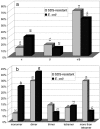
Most proteins are in equilibrium with partially and globally unfolded conformations. In contrast, kinetically stable proteins (KSPs) are trapped by an energy barrier in a specific state, unable to transiently sample other conformations. Among many potential roles, it appears that kinetic stability (KS) is a feature used by nature to allow proteins to maintain activity under harsh conditions and to preserve the structure of proteins that are prone to misfolding. The biological and pathological significance of KS remains poorly understood because of the lack of simple experimental methods to identify this property and its infrequent occurrence in proteins. Based on our previous correlation between KS and a protein's resistance to the denaturing detergent SDS, we show here the application of a diagonal 2D (D2D) SDS/PAGE assay to identify KSPs in complex mixtures. We applied this method to the lysate of Escherichia coli and upon proteomics analysis have identified 50 nonredundant proteins that were SDS-resistant (i.e., kinetically stable). Structural and functional analyses of a subset (44) of these proteins with known 3D structure revealed some potential structural and functional biases toward and against KS. This simple D2D SDS/PAGE assay will allow the widespread investigation of KS, including the proteomics-level identification of KSPs in different systems, potentially leading to a better understanding of the biological and pathological significance of this intriguing property of proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Plaza del Pino IM, Ibarra-Molero B, Sanchez-Ruiz JM. Proteins. 2000;40:58–70. - PubMed
-
- Sohl JL, Jaswal SS, Agard DA. Nature. 1998;395:817–819. - PubMed
-
- Kelly JW, Colón W, Lai Z, Lashuel HA, McCulloch J, McCutchen SL, Miroy GJ, Peterson SA. Adv Protein Chem. 1997;50:161–181. - PubMed
-
- Manning M, Colón W. Biochemistry. 2004;43:11248–11254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

